1EFG
 
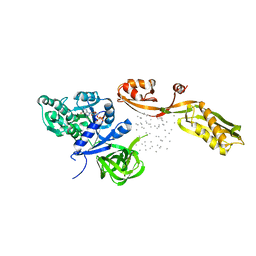 | | THE CRYSTAL STRUCTURE OF ELONGATION FACTOR G COMPLEXED WITH GDP, AT 2.7 ANGSTROMS RESOLUTION | | Descriptor: | ELONGATION FACTOR G, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Czworkowski, J, Wang, J, Steitz, T.A, Moore, P.B. | | Deposit date: | 1994-10-17 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of elongation factor G complexed with GDP, at 2.7 A resolution.
EMBO J., 13, 1994
|
|
1JJ2
 
 | | Fully Refined Crystal Structure of the Haloarcula marismortui Large Ribosomal Subunit at 2.4 Angstrom Resolution | | Descriptor: | 23S RRNA, 5S RRNA, CADMIUM ION, ... | | Authors: | Klein, D.J, Schmeing, T.M, Moore, P.B, Steitz, T.A. | | Deposit date: | 2001-07-03 | | Release date: | 2001-08-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The kink-turn: a new RNA secondary structure motif.
EMBO J., 20, 2001
|
|
1YJN
 
 | | Crystal Structure Of Clindamycin Bound To The G2099A Mutant 50S Ribosomal Subunit Of Haloarcula Marismortui | | Descriptor: | 23S Ribosomal RNA, 50S RIBOSOMAL PROTEIN L10E, 50S RIBOSOMAL PROTEIN L11P, ... | | Authors: | Tu, D, Blaha, G, Moore, P.B, Steitz, T.A. | | Deposit date: | 2005-01-14 | | Release date: | 2005-04-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance.
Cell(Cambridge,Mass.), 121, 2005
|
|
1YJ9
 
 | | Crystal Structure Of The Mutant 50S Ribosomal Subunit Of Haloarcula Marismortui Containing a three residue deletion in L22 | | Descriptor: | 23S Ribosomal RNA, 50S RIBOSOMAL PROTEIN L10E, 50S RIBOSOMAL PROTEIN L11P, ... | | Authors: | Tu, D, Blaha, G, Moore, P.B, Steitz, T.A. | | Deposit date: | 2005-01-13 | | Release date: | 2005-04-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance.
Cell(Cambridge,Mass.), 121, 2005
|
|
1YJW
 
 | | Crystal Structure Of Quinupristin Bound To The G2099A Mutant 50S Ribosomal Subunit Of Haloarcula Marismortui | | Descriptor: | 23S RIBOSOMAL RNA, 50S ribosomal protein L10, 50S ribosomal protein L10e, ... | | Authors: | Tu, D, Blaha, G, Moore, P.B, Steitz, T.A. | | Deposit date: | 2005-01-15 | | Release date: | 2005-04-26 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of Mlsbk Antibiotics Bound to Mutated Large Ribosomal Subunits Provide a Structural Explanation for Resistance.
Cell(Cambridge,Mass.), 121, 2005
|
|
2A9L
 
 | |
1U2A
 
 | |
3CPW
 
 | | The structure of the antibiotic LINEZOLID bound to the large ribosomal subunit of HALOARCULA MARISMORTUI | | Descriptor: | 23S RIBOSOMAL RNA, 5'-R(*CP*CP*AP*(PHE)*(ACA))-3', 50S ribosomal protein L10E, ... | | Authors: | Ippolito, J.A, Kanyo, Z.K, Wang, D, Franceschi, F.J, Moore, P.B, Steitz, T.A, Duffy, E.M. | | Deposit date: | 2008-04-01 | | Release date: | 2008-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the Oxazolidinone Antibiotic
Linezolid Bound to the 50S Ribosomal Subunit
J.Med.Chem., 51, 2008
|
|
1WTS
 
 | |
1WTT
 
 | |
1YI2
 
 | | Crystal Structure Of Erythromycin Bound To The G2099A Mutant 50S Ribosomal Subunit Of Haloarcula Marismortui | | Descriptor: | 23S Ribosomal RNA, 50S RIBOSOMAL PROTEIN L10E, 50S RIBOSOMAL PROTEIN L11P, ... | | Authors: | Tu, D, Blaha, G, Moore, P.B, Steitz, T.A. | | Deposit date: | 2005-01-11 | | Release date: | 2005-04-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance.
Cell(Cambridge,Mass.), 121, 2005
|
|
1YHQ
 
 | | Crystal Structure Of Azithromycin Bound To The G2099A Mutant 50S Ribosomal Subunit Of Haloarcula Marismortui | | Descriptor: | 23S Ribosomal RNA, 50S RIBOSOMAL PROTEIN L10E, 50S RIBOSOMAL PROTEIN L11P, ... | | Authors: | Tu, D, Blaha, G, Moore, P.B, Steitz, T.A. | | Deposit date: | 2005-01-10 | | Release date: | 2005-04-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance.
Cell(Cambridge,Mass.), 121, 2005
|
|
1YIT
 
 | | Crystal Structure Of Virginiamycin M and S Bound To The 50S Ribosomal Subunit Of Haloarcula Marismortui | | Descriptor: | 23S RIBOSOMAL RNA, 50S RIBOSOMAL PROTEIN L10E, 50S RIBOSOMAL PROTEIN L11P, ... | | Authors: | Tu, D, Blaha, G, Moore, P.B, Steitz, T.A. | | Deposit date: | 2005-01-13 | | Release date: | 2005-04-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of Mlsbk Antibiotics Bound to Mutated Large Ribosomal Subunits Provide a Structural Explanation for Resistance.
Cell(Cambridge,Mass.), 121, 2005
|
|
1YIJ
 
 | | Crystal Structure Of Telithromycin Bound To The G2099A Mutant 50S Ribosomal Subunit Of Haloarcula Marismortui | | Descriptor: | 23S Ribosomal RNA, 50S RIBOSOMAL PROTEIN L10E, 50S RIBOSOMAL PROTEIN L11P, ... | | Authors: | Tu, D, Blaha, G, Moore, P.B, Steitz, T.A. | | Deposit date: | 2005-01-12 | | Release date: | 2005-04-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance.
Cell(Cambridge,Mass.), 121, 2005
|
|
1JUR
 
 | |
1MHK
 
 | | Crystal Structure Analysis of a 26mer RNA molecule, representing a new RNA motif, the hook-turn | | Descriptor: | BROMIDE ION, RNA 12-mer BCh12, RNA 14-mer BCh12 | | Authors: | Szep, S, Wang, J, Moore, P.B. | | Deposit date: | 2002-08-20 | | Release date: | 2002-09-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of a 26-nucleotide RNA containing a hook-turn
RNA, 9, 2003
|
|
1KD1
 
 | | Co-crystal Structure of Spiramycin bound to the 50S Ribosomal Subunit of Haloarcula marismortui | | Descriptor: | 23S RRNA, 5S RRNA, CADMIUM ION, ... | | Authors: | Hansen, J.L, Ippolito, J.A, Ban, N, Nissen, P, Moore, P.B, Steitz, T.A. | | Deposit date: | 2001-11-12 | | Release date: | 2002-07-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structures of four macrolide antibiotics bound to the large ribosomal subunit.
Mol.Cell, 10, 2002
|
|
1M90
 
 | | Co-crystal structure of CCA-Phe-caproic acid-biotin and sparsomycin bound to the 50S ribosomal subunit | | Descriptor: | 23S RRNA, 5S RRNA, 6-AMINOHEXANOIC ACID, ... | | Authors: | Hansen, J.L, Schmeing, T.M, Moore, P.B, Steitz, T.A. | | Deposit date: | 2002-07-26 | | Release date: | 2002-09-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into peptide bond formation.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1M1K
 
 | | Co-crystal structure of azithromycin bound to the 50S ribosomal subunit of Haloarcula marismortui | | Descriptor: | 23S RRNA, 5S RRNA, AZITHROMYCIN, ... | | Authors: | Hansen, J.L, Ippolito, J.A, Ban, N, Nissen, P, Moore, P.B, Steitz, T.A. | | Deposit date: | 2002-06-19 | | Release date: | 2002-07-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structures of four macrolide antibiotics bound to the large ribosomal subunit.
Mol.Cell, 10, 2002
|
|
1OVY
 
 | |
1C04
 
 | | IDENTIFICATION OF KNOWN PROTEIN AND RNA STRUCTURES IN A 5 A MAP OF THE LARGE RIBOSOMAL SUBUNIT FROM HALOARCULA MARISMORTUI | | Descriptor: | 23S RRNA FRAGMENT, RIBOSOMAL PROTEIN L11, RIBOSOMAL PROTEIN L14, ... | | Authors: | Ban, N, Nissen, P, Capel, M, Moore, P.B, Steitz, T.A. | | Deposit date: | 1999-07-14 | | Release date: | 1999-08-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (5 Å) | | Cite: | Placement of protein and RNA structures into a 5 A-resolution map of the 50S ribosomal subunit.
Nature, 400, 1999
|
|
1SCL
 
 | |
1SZY
 
 | |
1A51
 
 | | LOOP D/LOOP E ARM OF E. COLI 5S RRNA, NMR, 9 STRUCTURES | | Descriptor: | 5S RRNA LOOP D/LOOP E | | Authors: | Dallas, A, Moore, P.B. | | Deposit date: | 1998-02-19 | | Release date: | 1998-05-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The loop E-loop D region of Escherichia coli 5S rRNA: the solution structure reveals an unusual loop that may be important for binding ribosomal proteins.
Structure, 5, 1997
|
|
1A4D
 
 | | LOOP D/LOOP E ARM OF ESCHERICHIA COLI 5S RRNA, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | RNA (5'-R(*GP*GP*CP*CP*GP*AP*UP*GP*GP*UP*AP*GP*UP*GP*UP*GP*GP*GP*GP*UP*C)-3'), RNA (5'-R(P*UP*CP*CP*CP*CP*AP*UP*GP*CP*GP*AP*GP*AP*GP*UP*AP*GP*GP*CP*C)-3') | | Authors: | Dallas, A, Moore, P.B. | | Deposit date: | 1998-01-29 | | Release date: | 1998-04-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The loop E-loop D region of Escherichia coli 5S rRNA: the solution structure reveals an unusual loop that may be important for binding ribosomal proteins.
Structure, 5, 1997
|
|
