1UGL
 
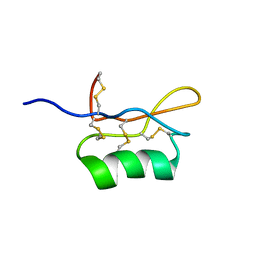 | | Solution structure of S8-SP11 | | Descriptor: | S-locus pollen protein | | Authors: | Mishima, M, Takayama, S, Sasaki, K, Jee, J.G, Kojima, C, Isogai, A, Shirakawa, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-16 | | Release date: | 2003-09-30 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structure of the Male Determinant Factor for Brassica Self-incompatibility
J.Biol.Chem., 278, 2003
|
|
1IRY
 
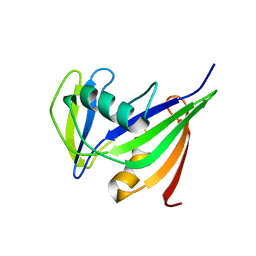 | | Solution structure of the hMTH1, a nucleotide pool sanitization enzyme | | Descriptor: | hMTH1 | | Authors: | Mishima, M, Itoh, N, Sakai, Y, Kamiya, H, Nakabeppu, Y, Shirakawa, M. | | Deposit date: | 2001-10-25 | | Release date: | 2003-12-23 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of human MTH1, a Nudix family hydrolase that selectively degrades oxidized purine nucleoside triphosphates
J.Biol.Chem., 279, 2004
|
|
1X60
 
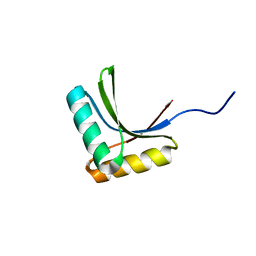 | | Solution structure of the peptidoglycan binding domain of B. subtilis cell wall lytic enzyme CwlC | | Descriptor: | Sporulation-specific N-acetylmuramoyl-L-alanine amidase | | Authors: | Mishima, M, Shida, T, Yabuki, K, Kato, K, Sekiguchi, J, Kojima, C. | | Deposit date: | 2005-05-17 | | Release date: | 2005-08-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Peptidoglycan Binding Domain of Bacillus subtilis Cell Wall Lytic Enzyme CwlC: Characterization of the Sporulation-Related Repeats by NMR(,)
Biochemistry, 44, 2005
|
|
2E4H
 
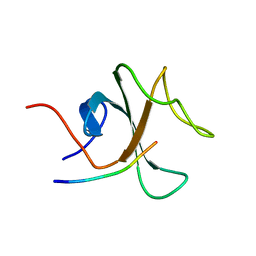 | |
2E30
 
 | |
7Y4N
 
 | | Insight into the C-terminal SH3 domain mediated binding of Drosophila Drk to Sos and Dos | | Descriptor: | Growth factor receptor-bound protein 2 | | Authors: | Pooppadi, M.S, Ikeya, T, Sugasawa, H, Watanabe, R, Mishima, M, Inomata, K, Ito, Y. | | Deposit date: | 2022-06-15 | | Release date: | 2022-08-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Insight into the C-terminal SH3 domain mediated binding of Drosophila Drk to Sos and Dos.
Biochem.Biophys.Res.Commun., 625, 2022
|
|
8K9O
 
 | |
8COG
 
 | | Human arginylated beta-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Pinto, C.S, Bakker, S.E, Suchenko, A, Hussain, H, Hatano, T, Sampath, K, Chinthalapudi, K, Mishima, M, Balasubramanian, M. | | Deposit date: | 2023-02-28 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.499 Å) | | Cite: | Structure and physiological investigation of human arginylated beta-actin
To Be Published
|
|
2N9K
 
 | | 1H, 13C, and 15N Chemical Shift Assignments for in vitro GB1 | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Ikeya, T, Hanashima, T, Hosoya, S, Shimazaki, M, Ikeda, S, Mishima, M, Guentert, P, Ito, Y. | | Deposit date: | 2015-11-26 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Improved in-cell structure determination of proteins at near-physiological concentration
Sci Rep, 6, 2016
|
|
2N9L
 
 | | 1H, 13C, and 15N Chemical Shift Assignments for in-cell GB1 | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Ikeya, T, Hanashima, T, Hosoya, S, Shimazaki, M, Ikeda, S, Mishima, M, Guentert, P, Ito, Y. | | Deposit date: | 2015-11-30 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Improved in-cell structure determination of proteins at near-physiological concentration
Sci Rep, 6, 2016
|
|
1J0S
 
 | | Solution structure of the human interleukin-18 | | Descriptor: | Interleukin-18 | | Authors: | Kato, Z, Jee, J, Shikano, H, Mishima, M, Ohki, I, Yoneda, T, Hara, T, Torigoe, K, Kondo, N, Shirakawa, M. | | Deposit date: | 2002-11-21 | | Release date: | 2003-11-11 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The structure and binding mode of interleukin-18
Nat.Struct.Biol., 10, 2003
|
|
7CKV
 
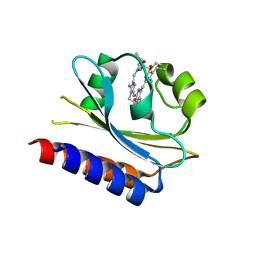 | | Crystal structure of Cyanobacteriochrome GAF domain in Pr state | | Descriptor: | MAGNESIUM ION, PHYCOCYANOBILIN, RcaE | | Authors: | Nagae, T, Koizumi, T, Hirose, Y, Mishima, M. | | Deposit date: | 2020-07-19 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural basis of the protochromic green/red photocycle of the chromatic acclimation sensor RcaE.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1X26
 
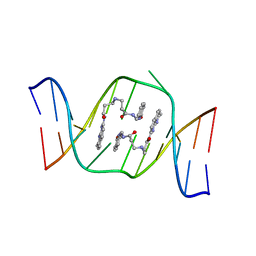 | | Solution structure of the AA-mismatch DNA complexed with naphthyridine-azaquinolone | | Descriptor: | 5'-D(*CP*AP*TP*TP*CP*AP*GP*TP*TP*AP*G)-3', 5'-D(*CP*TP*AP*AP*CP*AP*GP*AP*AP*TP*G)-3', N~3~-{3-[(7-METHYL-1,8-NAPHTHYRIDIN-2-YL)AMINO]-3-OXOPROPYL}-N~1~-[(7-OXO-7,8-DIHYDRO-1,8-NAPHTHYRIDIN-2-YL)METHYL]-BET A-ALANINAMIDE | | Authors: | Nakatani, K, Hagihara, S, Goto, Y, Kobori, A, Hagihara, M, Hayashi, G, Kyo, M, Nomura, M, Mishima, M, Kojima, C. | | Deposit date: | 2005-04-20 | | Release date: | 2006-04-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Small-molecule ligand induces nucleotide flipping in (CAG)n trinucleotide repeats
Nat.Chem.Biol., 1, 2005
|
|
5ZD0
 
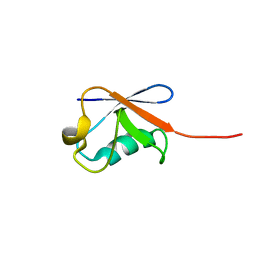 | | Solution structure of human ubiquitin with three alanine mutations in living eukaryotic cells by in-cell NMR spectroscopy | | Descriptor: | ubiquitin | | Authors: | Tanaka, T, Ikeya, T, Kamoshida, H, Mishima, M, Shirakawa, M, Guentert, P, Ito, Y. | | Deposit date: | 2018-02-22 | | Release date: | 2019-08-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Protein 3D Structure Determination in Living Eukaryotic Cells.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
5Z4B
 
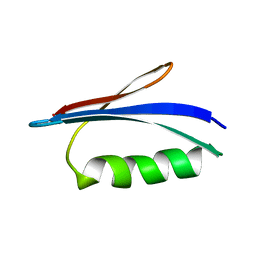 | | GB1 structure determination in living eukaryotic cells by in-cell NMR spectroscopy | | Descriptor: | Protein LG | | Authors: | Tanaka, T, Teppei, I, Kamoshida, H, Mishima, M, Shirakawa, M, Guentert, P, Ito, Y. | | Deposit date: | 2018-01-10 | | Release date: | 2019-01-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Protein 3D Structure Determination in Living Eukaryotic Cells.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
5ZCZ
 
 | | Solution structure of the T. Thermophilus HB8 TTHA1718 protein in living eukaryotic cells by in-cell NMR spectroscopy | | Descriptor: | Heavy metal binding protein | | Authors: | Tanaka, T, Teppei, I, Kamoshida, H, Mishima, M, Shirakawa, M, Guentert, P, Ito, Y. | | Deposit date: | 2018-02-22 | | Release date: | 2019-08-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Protein 3D Structure Determination in Living Eukaryotic Cells.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
2ROG
 
 | | Solution structure of Thermus thermophilus HB8 TTHA1718 protein in living E. coli cells | | Descriptor: | Heavy metal binding protein | | Authors: | Sakakibara, D, Sasaki, A, Ikeya, T, Hamatsu, J, Koyama, H, Mishima, M, Mikawa, T, Waelchli, M, Smith, B.O, Shirakawa, M, Guentert, P, Ito, Y. | | Deposit date: | 2008-03-21 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination in living cells by in-cell NMR spectroscopy
Nature, 458, 2009
|
|
2ROE
 
 | | Solution structure of thermus thermophilus HB8 TTHA1718 protein in vitro | | Descriptor: | Heavy metal binding protein | | Authors: | Sakakibara, D, Sasaki, A, Ikeya, T, Hamatsu, J, Koyama, H, Mishima, M, Mikawa, T, Waelchli, M, Smith, B.O, Shirakawa, M, Guentert, P, Ito, Y. | | Deposit date: | 2008-03-20 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination in living cells by in-cell NMR spectroscopy
Nature, 458, 2009
|
|
2RQL
 
 | |
2RRD
 
 | | Structure of HRDC domain from human Bloom syndrome protein, BLM | | Descriptor: | HRDC domain from Bloom syndrome protein | | Authors: | Sato, A, Mishima, M, Nagai, A, Kim, S.Y, Ito, Y, Hakoshima, T, Jee, J.G, Kitano, K. | | Deposit date: | 2010-07-19 | | Release date: | 2010-09-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the HRDC domain of human Bloom syndrome protein BLM
J.Biochem., 148, 2010
|
|
2RR3
 
 | | Solution structure of the complex between human VAP-A MSP domain and human OSBP FFAT motif | | Descriptor: | Oxysterol-binding protein 1, Vesicle-associated membrane protein-associated protein A | | Authors: | Furuita, K, Jee, J, Fukada, H, Mishima, M, Kojima, C. | | Deposit date: | 2010-03-09 | | Release date: | 2010-03-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Electrostatic interaction between oxysterol-binding protein and VAMP-associated protein A revealed by NMR and mutagenesis studies
J.Biol.Chem., 285, 2010
|
|
2RRK
 
 | |
2RT5
 
 | |
2RM8
 
 | | The solution structure of phototactic transducer protein HtrII linker region from Natronomonas pharaonis | | Descriptor: | Sensory rhodopsin II transducer | | Authors: | Hayashi, K, Sudo, Y, Jee, J, Mishima, M, Hara, H, Kamo, N, Kojima, C. | | Deposit date: | 2007-10-15 | | Release date: | 2007-12-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of the Phototactic Transducer Protein HtrII Linker Region from Natronomonas pharaonis
Biochemistry, 46, 2007
|
|
2RT3
 
 | | Solution structure of the second RRM domain of Nrd1 | | Descriptor: | Negative regulator of differentiation 1 | | Authors: | Kobayashi, A, Kanaba, T, Mishima, M. | | Deposit date: | 2013-04-16 | | Release date: | 2014-04-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the second RRM domain of Nrd1, a fission yeast MAPK target RNA binding protein, and implication for its RNA recognition and regulation
Biochem.Biophys.Res.Commun., 437, 2013
|
|
