3CKP
 
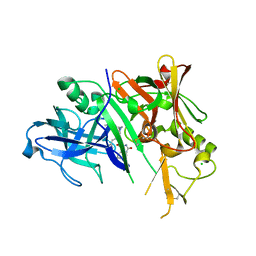 | | Crystal structure of BACE-1 in complex with inhibitor | | Descriptor: | (4S)-N-[(1S,2R)-1-benzyl-3-{[3-(dimethylamino)benzyl]amino}-2-hydroxypropyl]-1-(3-methoxybenzyl)-2-oxoimidazolidine-4-carboxamide, Beta-secretase 1, CHLORIDE ION | | Authors: | Min, K. | | Deposit date: | 2008-03-16 | | Release date: | 2008-06-03 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Synthesis, SAR, and X-ray structure of human BACE-1 inhibitors with cyclic urea derivatives
Bioorg.Med.Chem.Lett., 18, 2008
|
|
1PCV
 
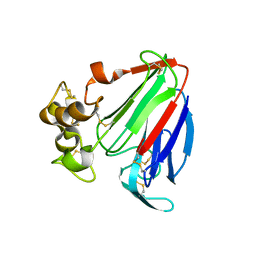 | | Crystal structure of osmotin, a plant antifungal protein | | Descriptor: | osmotin | | Authors: | Min, K, Ha, S.C, Yun, D.-J, Bressan, R.A, Kim, K.K. | | Deposit date: | 2003-05-16 | | Release date: | 2004-02-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of osmotin, a plant antifungal protein
PROTEINS: STRUCT.,FUNCT.,GENET., 54, 2004
|
|
3ENQ
 
 | |
3ENW
 
 | |
3ENV
 
 | |
3CKR
 
 | | Crystal structure of BACE-1 in complex with inhibitor | | Descriptor: | (4S)-1,4-dibenzyl-N-[(1S,2R)-1-benzyl-3-{[3-(dimethylamino)benzyl]amino}-2-hydroxypropyl]-2-oxoimidazolidine-4-carboxamide, Beta-secretase 1 | | Authors: | Min, K. | | Deposit date: | 2008-03-16 | | Release date: | 2008-06-03 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Synthesis, SAR, and X-ray structure of human BACE-1 inhibitors with cyclic urea derivatives
Bioorg.Med.Chem.Lett., 18, 2008
|
|
7E60
 
 | | The crystal structure of peptidoglycan peptidase in complex with inhibitor 1 | | Descriptor: | (2~{R},6~{S})-2,6-diacetamido-7-[[(2~{R})-1-(oxidanylamino)-1-oxidanylidene-propan-2-yl]amino]-7-oxidanylidene-heptanoic acid, Peptidase M23, ZINC ION | | Authors: | Min, K, Yoon, H.J, Choi, Y, Lee, H.H. | | Deposit date: | 2021-02-21 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure-based inhibitor design for reshaping bacterial morphology
Commun Biol, 5, 2022
|
|
1JXV
 
 | | Crystal Structure of Human Nucleoside Diphosphate Kinase A | | Descriptor: | Nucleoside Diphosphate Kinase A | | Authors: | Min, K, Song, H.K, Chang, C, Kim, S.Y, Lee, K.J, Suh, S.W. | | Deposit date: | 2001-09-10 | | Release date: | 2002-04-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human nucleoside diphosphate kinase A, a metastasis suppressor.
Proteins, 46, 2002
|
|
1EK8
 
 | |
2LCX
 
 | |
2L2T
 
 | |
2L9U
 
 | |
2L03
 
 | |
6XYI
 
 | | NMR solution structure of alpha-AnmTX- Ms11a-3 (Ms11a-3) | | Descriptor: | AMS9.3.1 | | Authors: | Mineev, K.S, Kornilov, F.D, Lushpa, V.A, Logashina, Y.A, Maleeva, E.E, Andreev, Y.A. | | Deposit date: | 2020-01-30 | | Release date: | 2021-02-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of alpha-AnmTX- Ms11a-3 (Ms11a-3)
TO BE PUBLISHED
|
|
6XYH
 
 | | NMR solution structure of alpha-AnmTX-Ms11a-2 (Ms11a-2) | | Descriptor: | AMS3 | | Authors: | Mineev, K.S, Kornilov, F.D, Lushpa, V.A, Logashina, Y.A, Maleeva, E.E, Andreev, Y.A. | | Deposit date: | 2020-01-30 | | Release date: | 2021-02-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of alpha-AnmTX-Ms11a-2 (Ms11a-2)
To Be Published
|
|
5LAH
 
 | | NMR structure of the sea anemone peptide tau-AnmTx Ueq 12-1 with an uncommon fold | | Descriptor: | tau-AnmTx Ueq 12-1 | | Authors: | Mineev, K.S, Arseniev, A.S, Andreev, Y.A, Kozlov, S.A, Logashina, Y.A. | | Deposit date: | 2016-06-14 | | Release date: | 2017-05-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | New Disulfide-Stabilized Fold Provides Sea Anemone Peptide to Exhibit Both Antimicrobial and TRPA1 Potentiating Properties
Toxins, 9, 2017
|
|
7NT7
 
 | |
5LQV
 
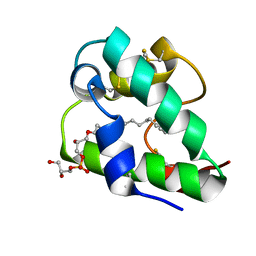 | | Spatial structure of the lentil lipid transfer protein in complex with anionic lysolipid LPPG | | Descriptor: | 1-MYRISTOYL-2-HYDROXY-SN-GLYCERO-3-[PHOSPHO-RAC-(1-GLYCEROL)], Non-specific lipid-transfer protein 2 | | Authors: | Mineev, K.S, Shenkarev, Z.O, Arseniev, A.S, Melnikova, D.N, Finkina, E.I, Ovchinnikova, T.V. | | Deposit date: | 2016-08-17 | | Release date: | 2017-06-28 | | Last modified: | 2019-05-08 | | Method: | SOLUTION NMR | | Cite: | Ligand Binding Properties of the Lentil Lipid Transfer Protein: Molecular Insight into the Possible Mechanism of Lipid Uptake.
Biochemistry, 56, 2017
|
|
5LM0
 
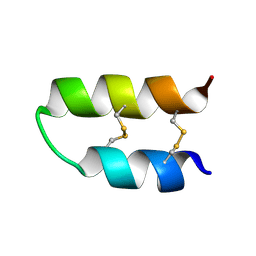 | | NMR spatial structure of Tk-hefu peptide | | Descriptor: | L-2 | | Authors: | Mineev, K.S, Berkut, A.A, Novikova, E.V, Oparin, P.B, Grishin, E.V, Arseniev, A.S, Vassilevski, A.A. | | Deposit date: | 2016-07-28 | | Release date: | 2017-08-16 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Protein Surface Topography as a tool to enhance the selective activity of a potassium channel blocker.
J.Biol.Chem., 2019
|
|
5MOU
 
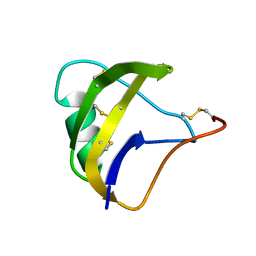 | | NMR spatial structure of scorpion alpha-like toxin BeM9 | | Descriptor: | Alpha-mammal toxin BeM9 | | Authors: | Mineev, K.S, Kuldushev, N.A, Berkut, A.A, Grishin, E.V, Vassilevski, A.A, Arseniev, A.S. | | Deposit date: | 2016-12-14 | | Release date: | 2018-01-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Refined structure of BeM9 reveals arginine hand, an overlooked structural motif in scorpion toxins affecting sodium channels.
Proteins, 86, 2018
|
|
6THI
 
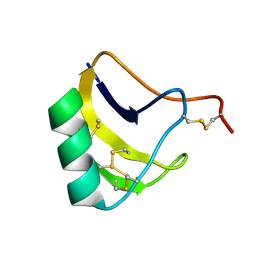 | | Solution structure of MeuNaTxalpha-1 toxin from Mesobuthus Eupeus | | Descriptor: | Sodium channel neurotoxin MeuNaTxalpha-1 | | Authors: | Mineev, K.S, Kuzmenkov, A.I, Khusainov, G.A, Arseniev, A.S, Vassilevski, A.A. | | Deposit date: | 2019-11-20 | | Release date: | 2020-12-02 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure of MeuNaTx alpha-1 toxin from scorpion venom highlights the importance of the nest motif.
Proteins, 2021
|
|
7AVB
 
 | |
7AVA
 
 | |
7AY8
 
 | | NMR solution structure of Tbo-IT2 | | Descriptor: | Tbo-IT2 | | Authors: | Mineev, K.S, Kornilov, F.D, Lushpa, V.A, Korolkova, Y.A, Maleeva, E.E, Andreev, Y.A, Kozlov, S.A, Arseniev, A.S. | | Deposit date: | 2020-11-11 | | Release date: | 2021-03-03 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | New Insectotoxin from Tibellus Oblongus Spider Venom Presents Novel Adaptation of ICK Fold.
Toxins, 13, 2021
|
|
6GWM
 
 | | Solution structure of rat RIP2 caspase recruitment domain | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 2 | | Authors: | Mineev, K.S, Goncharuk, S.A, Artemieva, L.E, Arseniev, A.S. | | Deposit date: | 2018-06-25 | | Release date: | 2018-10-31 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | CARD domain of rat RIP2 kinase: Refolding, solution structure, pH-dependent behavior and protein-protein interactions.
PLoS ONE, 13, 2018
|
|
