3HR8
 
 | | Crystal Structure of Thermotoga maritima RecA | | Descriptor: | Protein recA | | Authors: | Lee, S, Kim, T.G, Jeong, E.-Y, Ban, C, Jeon, W.-J, Min, K.I, Song, K.-M, Heo, S.-D, Ku, J.K. | | Deposit date: | 2009-06-09 | | Release date: | 2010-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of RecA Protein from Thermotoga maritima MSB8
to be published
|
|
4NXZ
 
 | | DNA polymerase beta with O6mG in the template base opposite to incoming non-hydrolyzable TTP with manganese in the active site | | Descriptor: | 5'-D(*CP*CP*GP*AP*CP*(6OG)P*TP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*A)-3', 5'-D(P*GP*TP*CP*GP*G)-3', ... | | Authors: | Koag, M.-C, Min, K, Monzingo, A.F, Lee, S. | | Deposit date: | 2013-12-09 | | Release date: | 2014-04-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.557 Å) | | Cite: | Metal-dependent conformational activation explains highly promutagenic replication across O6-methylguanine by human DNA polymerase beta.
J.Am.Chem.Soc., 136, 2014
|
|
4NY8
 
 | | DNA polymerase beta with O6mG in the template base opposite to incoming non-hydrolyzable CTP with manganese in the active site | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, 5'-D(*CP*CP*GP*AP*CP*(6OG)P*TP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*A)-3', ... | | Authors: | Koag, M.-C, Min, K, Monzingo, A.F, Lee, S. | | Deposit date: | 2013-12-10 | | Release date: | 2014-04-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.246 Å) | | Cite: | Metal-dependent conformational activation explains highly promutagenic replication across O6-methylguanine by human DNA polymerase beta.
J.Am.Chem.Soc., 136, 2014
|
|
4MFC
 
 | | Structure of human DNA polymerase beta complexed with O6MG in the template base paired with incoming non-hydrolyzable CTP | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA polymerase beta, MAGNESIUM ION, ... | | Authors: | Koag, M.C, Min, K, Monzingo, A.F, Lee, S. | | Deposit date: | 2013-08-27 | | Release date: | 2014-04-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Metal-dependent conformational activation explains highly promutagenic replication across O6-methylguanine by human DNA polymerase beta.
J.Am.Chem.Soc., 136, 2014
|
|
4MFF
 
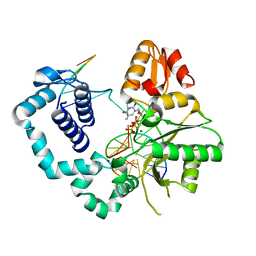 | | Structure of human DNA polymerase beta complexed with O6MG in the template base paired with incoming non-hydrolyzable TTP | | Descriptor: | 5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]thymidine, DNA polymerase beta, MAGNESIUM ION, ... | | Authors: | Koag, M.C, Min, K, Monzingo, A.F, Lee, S. | | Deposit date: | 2013-08-27 | | Release date: | 2014-04-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Metal-dependent conformational activation explains highly promutagenic replication across O6-methylguanine by human DNA polymerase beta.
J.Am.Chem.Soc., 136, 2014
|
|
1RZL
 
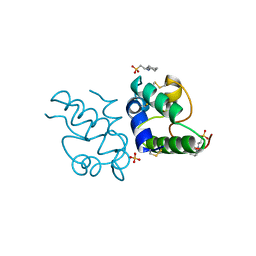 | | RICE NONSPECIFIC LIPID TRANSFER PROTEIN | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, NONSPECIFIC LIPID TRANSFER PROTEIN, SULFATE ION | | Authors: | Lee, J.Y, Min, K.S, Cha, H, Shin, D.H, Hwang, K.Y, Suh, S.W. | | Deposit date: | 1997-10-09 | | Release date: | 1998-12-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rice non-specific lipid transfer protein: the 1.6 A crystal structure in the unliganded state reveals a small hydrophobic cavity.
J.Mol.Biol., 276, 1998
|
|
4V0Q
 
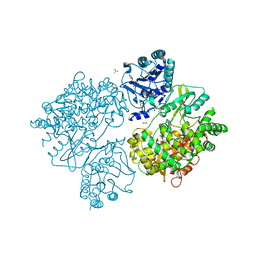 | | Dengue Virus Full Length NS5 Complexed with SAH | | Descriptor: | ACETATE ION, GLYCEROL, NS5 POLYMERASE, ... | | Authors: | Zhao, Y, Soh, S, Zheng, J, Phoo, W.W, Swaminathan, K, Cornvik, T.C, Lim, S.P, Shi, P.-Y, Lescar, J, Vasudevan, S.G, Luo, D. | | Deposit date: | 2014-09-18 | | Release date: | 2015-01-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Crystal Structure of the Dengue Virus Ns5 Protein Reveals a Novel Inter-Domain Interface Essential for Protein Flexibility and Virus Replication.
Plos Pathog., 11, 2015
|
|
4URG
 
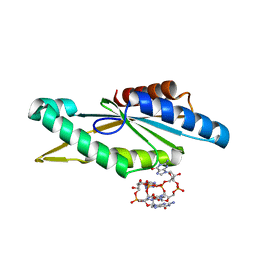 | | Crystal Structure of GGDEF domain from T.maritima (active-like dimer) | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), DIGUANYLATE CYCLASE | | Authors: | Deepthi, A, Liew, C.W, Liang, Z.X, Swaminathan, K, Lescar, J. | | Deposit date: | 2014-06-30 | | Release date: | 2014-10-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a Diguanylate Cyclase from Thermotoga Maritima: Insights Into Activation, Feedback Inhibition and Thermostability
Plos One, 9, 2014
|
|
7AJP
 
 | | Crystal Structure of Human Adenovirus 56 Fiber Knob | | Descriptor: | 1,2-ETHANEDIOL, Fiber, GLYCEROL, ... | | Authors: | Strebl, M, Mindler, K, Stehle, T. | | Deposit date: | 2020-09-29 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.382018 Å) | | Cite: | Human species D adenovirus hexon capsid protein mediates cell entry through a direct interaction with CD46.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8AO1
 
 | | solution structure of nanoFAST fluorogen-activating protein in the apo state | | Descriptor: | nanoFAST | | Authors: | Lushpa, V.A, Goncharuk, M.V, Goncharuk, S.A, Baranov, M.S, Mineev, K.S. | | Deposit date: | 2022-08-08 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Spatial Structure of NanoFAST in the Apo State and in Complex with its Fluorogen HBR-DOM2.
Int J Mol Sci, 23, 2022
|
|
8AO0
 
 | | Solution structure of nanoFAST/HBR-DOM2 complex | | Descriptor: | (5~{Z})-5-[(2,5-dimethoxy-4-oxidanyl-phenyl)methylidene]-2-sulfanylidene-1,3-thiazolidin-4-one, Photoactive yellow protein | | Authors: | Lushpa, V.A, Goncharuk, M.V, Goncharuk, S.A, Baleeva, N.S, Baranov, M.S, Mineev, K.S. | | Deposit date: | 2022-08-08 | | Release date: | 2022-11-23 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Spatial Structure of NanoFAST in the Apo State and in Complex with its Fluorogen HBR-DOM2.
Int J Mol Sci, 23, 2022
|
|
7AV6
 
 | | FAST in a domain-swapped dimer form | | Descriptor: | FORMIC ACID, Photoactive yellow protein | | Authors: | Bukhdruker, S, Remeeva, A, Ruchkin, D, Gorbachev, D, Povarova, N, Mineev, K, Goncharuk, S, Baranov, M, Mishin, A, Borshchevskiy, V. | | Deposit date: | 2020-11-04 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | NanoFAST: structure-based design of a small fluorogen-activating protein with only 98 amino acids.
Chem Sci, 12, 2021
|
|
8AR3
 
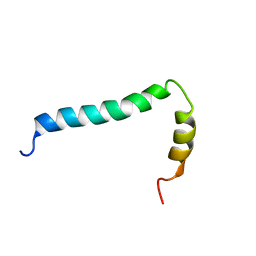 | | Solution structure of TLR9 transmembrane and cytoplasmic juxtamembrane regions | | Descriptor: | Toll-like receptor 9 | | Authors: | Kornilov, F.D, Shabalkina, A.V, Goncharuk, M.V, Goncharuk, S.A, Arseniev, A.S, Mineev, K.S. | | Deposit date: | 2022-08-15 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The architecture of transmembrane and cytoplasmic juxtamembrane regions of Toll-like receptors.
Nat Commun, 14, 2023
|
|
8AR1
 
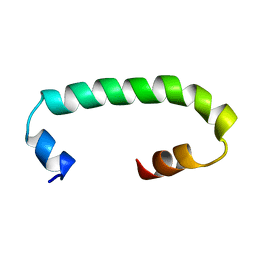 | | Solution structure of TLR3 transmembrane and cytoplasmic juxtamembrane regions | | Descriptor: | Toll-like receptor 3 | | Authors: | Kornilov, F.D, Shabalkina, A.V, Goncharuk, M.V, Goncharuk, S.A, Arseniev, A.S, Mineev, K.S. | | Deposit date: | 2022-08-15 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The architecture of transmembrane and cytoplasmic juxtamembrane regions of Toll-like receptors.
Nat Commun, 14, 2023
|
|
8AR0
 
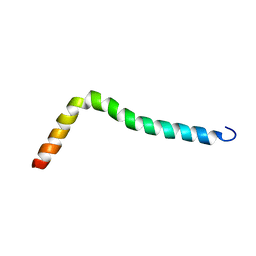 | | Solution structure of TLR2 transmembrane and cytoplasmic juxtamembrane regions | | Descriptor: | Toll-like receptor 2 | | Authors: | Kornilov, F.D, Shabalkina, A.V, Goncharuk, M.V, Goncharuk, S.A, Arseniev, A.S, Mineev, K.S. | | Deposit date: | 2022-08-15 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The architecture of transmembrane and cytoplasmic juxtamembrane regions of Toll-like receptors.
Nat Commun, 14, 2023
|
|
8AR2
 
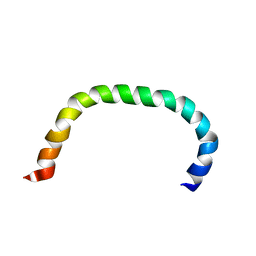 | | Solution structure of TLR5 transmembrane and cytoplasmic juxtamembrane regions | | Descriptor: | Toll-like receptor 5 | | Authors: | Shabalkina, A.V, Kornilov, F.D, Goncharuk, M.V, Goncharuk, S.A, Arseniev, A.S, Mineev, K.S. | | Deposit date: | 2022-08-15 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The architecture of transmembrane and cytoplasmic juxtamembrane regions of Toll-like receptors.
Nat Commun, 14, 2023
|
|
7FGJ
 
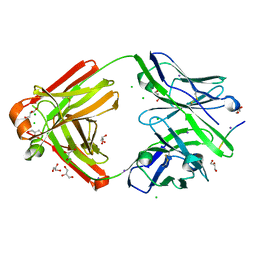 | | The cross-reaction complex structure with VQILNK peptide and the tau antibody's Fab domain. | | Descriptor: | CHLORIDE ION, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Tsuchida, T, Tsuchiya, T, Miyamoto, K, In, Y, Minoura, K, Taniguchi, T, Ishida, T, Tomoo, K. | | Deposit date: | 2021-07-27 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The cross-reaction complex structure with VQILNK peptide and the antibody's Fab domain.
To Be Published
|
|
7FGR
 
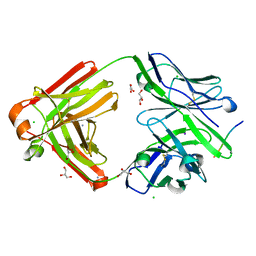 | | The cross-reaction complex structure with VQIFNK peptide and the tau antibody's Fab domain. | | Descriptor: | AMMONIUM ION, CHLORIDE ION, Fab Heavy Chain, ... | | Authors: | Tsuchida, T, Tsuchiya, T, Miyamoto, K, In, Y, Minoura, K, Taniguchi, T, Ishida, T, Tomoo, K. | | Deposit date: | 2021-07-27 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The cross-reaction complex structure with VQIFNK peptide and the tau antibody's Fab domain.
To Be Published
|
|
7FGK
 
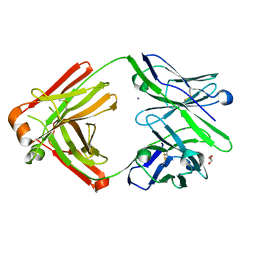 | | The Fab antibody single structure against tau protein. | | Descriptor: | Fab Heavy Chain, Fab Light Chain, GLYCEROL, ... | | Authors: | Tsuchida, T, Susa, K, Kibiki, T, Tsuchiya, T, Miyamoto, K, In, Y, Minoura, K, Taniguchi, T, Ishida, T, Tomoo, K. | | Deposit date: | 2021-07-27 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The free structure of the Fab domain of antibody that recognizes the PHF core region VQIINK in Tau protein.
To Be Published
|
|
6LKB
 
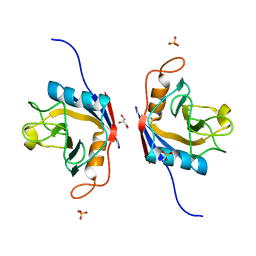 | | Crystal Structure of the peptidylprolyl isomerase domain of Arabidopsis thaliana CYP71. | | Descriptor: | COBALT (II) ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Lakhanpal, S, Jobichen, C, Swaminathan, K. | | Deposit date: | 2019-12-18 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Structural and functional analyses of the PPIase domain of Arabidopsis thaliana CYP71 reveal its catalytic activity toward histone H3.
Febs Lett., 595, 2021
|
|
3FHV
 
 | |
3FHU
 
 | |
8EVD
 
 | |
3BCD
 
 | | Alpha-amylase B in complex with maltotetraose and alpha-cyclodextrin | | Descriptor: | Alpha amylase, catalytic region, CALCIUM ION, ... | | Authors: | Tan, T.-C, Mijts, B.N, Swaminathan, K, Patel, B.K.C, Divne, C. | | Deposit date: | 2007-11-12 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Polyextremophilic alpha-Amylase AmyB from Halothermothrix orenii: Details of a Productive Enzyme-Substrate Complex and an N Domain with a Role in Binding Raw Starch
J.Mol.Biol., 378, 2008
|
|
5LV6
 
 | | N-terminal motif dimerization of EGFR transmembrane domain in bicellar environment | | Descriptor: | Epidermal growth factor receptor | | Authors: | Bragin, P, Bocharov, E, Mineev, K, Bocharova, O, Arseniev, A. | | Deposit date: | 2016-09-12 | | Release date: | 2017-04-05 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The Conformation of the Epidermal Growth Factor Receptor Transmembrane Domain Dimer Dynamically Adapts to the Local Membrane Environment.
Biochemistry, 56, 2017
|
|
