2O05
 
 | | Human spermidine synthase | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, Spermidine synthase | | Authors: | Min, J, Wu, H, Zeng, H, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-11-27 | | Release date: | 2006-12-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and mechanism of spermidine synthases.
Biochemistry, 46, 2007
|
|
2O0L
 
 | | Human spermidine synthase | | Descriptor: | 5'-[(S)-(3-AMINOPROPYL)(METHYL)-LAMBDA~4~-SULFANYL]-5'-DEOXYADENOSINE, Spermidine synthase | | Authors: | Min, J, Wu, H, Zeng, H, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-11-27 | | Release date: | 2006-12-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure and mechanism of spermidine synthases.
Biochemistry, 46, 2007
|
|
2QSH
 
 | | Crystal structure of Rad4-Rad23 bound to a mismatch DNA | | Descriptor: | DNA repair protein RAD4, UV excision repair protein RAD23, bottom strand of the mismatch DNA, ... | | Authors: | Min, J.-H, Pavletich, N.P. | | Deposit date: | 2007-07-31 | | Release date: | 2007-10-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | Recognition of DNA damage by the Rad4 nucleotide excision repair protein
Nature, 449, 2007
|
|
2QSG
 
 | |
2QSF
 
 | | Crystal structure of the Rad4-Rad23 complex | | Descriptor: | DNA repair protein RAD4, UV excision repair protein RAD23 | | Authors: | Min, J.-H, Pavletich, N.P. | | Deposit date: | 2007-07-31 | | Release date: | 2007-10-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Recognition of DNA damage by the Rad4 nucleotide excision repair protein
Nature, 449, 2007
|
|
2AVD
 
 | | Crystal Structure of Human Catechol-O-methyltransferase domain containing 1 | | Descriptor: | Catechol-O-methyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Min, J.R, Wu, H, Zeng, H, Loppnau, P, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-08-29 | | Release date: | 2005-09-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of Human Catechol-O-methyltransferase
domain containing 1 in complex with S-adenosyl-L-methionine
To be Published
|
|
2FYT
 
 | | Human HMT1 hnRNP methyltransferase-like 3 (S. cerevisiae) protein | | Descriptor: | Protein arginine N-methyltransferase 3, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Min, J.R, Wu, H, Zeng, H, Loppnau, P, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-02-08 | | Release date: | 2006-03-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of Human HMT1 hnRNP
methyltransferase-like 3 in complex with SAH.
To be Published
|
|
4Z5H
 
 | | HipB(S29A)-O2 20mer complex | | Descriptor: | Antitoxin HipB, DNA (5'-D(*TP*TP*AP*TP*CP*CP*TP*CP*AP*CP*TP*AP*AP*AP*GP*GP*AP*TP*AP*A)-3') | | Authors: | Min, J, Brennan, R.G, Schumacher, M.A. | | Deposit date: | 2015-04-02 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular mechanism on hipBA gene regulation.
To be published
|
|
4Z5C
 
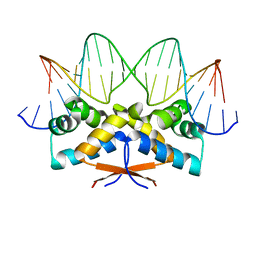 | | HipB-O3 21mer complex | | Descriptor: | Antitoxin HipB, DNA (5'-D(*TP*TP*TP*AP*TP*CP*CP*CP*GP*TP*AP*GP*AP*GP*CP*GP*GP*AP*TP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*AP*TP*CP*CP*GP*CP*TP*CP*TP*AP*CP*GP*GP*GP*AP*TP*A)-3') | | Authors: | Min, J, Brennan, R.G, Schumacher, M.A. | | Deposit date: | 2015-04-02 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mechanism on hipBA gene regulation.
To be published
|
|
4Z59
 
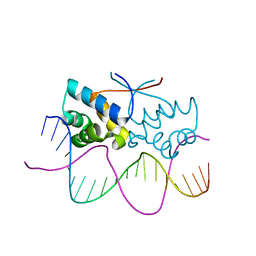 | | HipB-O4 20mer complex | | Descriptor: | Antitoxin HipB, DNA (5'-D(*TP*TP*AP*TP*CP*CP*GP*CP*GP*AP*TP*CP*GP*CP*GP*GP*AP*TP*AP*A)-3') | | Authors: | Min, J, Brennan, R.G, Schumacher, M.A. | | Deposit date: | 2015-04-02 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular mechanism on hipBA gene regulation.
To be published
|
|
4Z5D
 
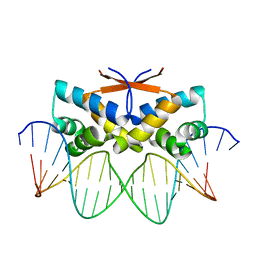 | | HipB-O4 21mer complex | | Descriptor: | Antitoxin HipB, DNA (5'-D(*TP*TP*TP*AP*TP*CP*CP*GP*CP*GP*AP*TP*CP*GP*CP*GP*GP*AP*TP*A)-3'), DNA (5'-D(*TP*TP*TP*AP*TP*CP*CP*GP*CP*GP*AP*TP*CP*GP*CP*GP*GP*AP*TP*AP*A)-3') | | Authors: | Min, J, Brennan, R.G, Schumacher, M.A. | | Deposit date: | 2015-04-02 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular mechanism on hipBA gene regulation.
To be published
|
|
4Z58
 
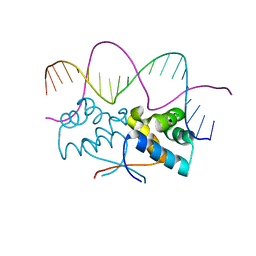 | | HipB-O3 20mer complex | | Descriptor: | Antitoxin HipB, DNA (5'-D(*TP*TP*AP*TP*CP*CP*GP*CP*TP*CP*TP*AP*CP*GP*GP*GP*AP*TP*AP*A)-3') | | Authors: | Min, J, Brennan, R.G, Schumacher, M.A. | | Deposit date: | 2015-04-02 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mechanism on hipBA gene regulation.
To be published
|
|
6WAT
 
 | | complex structure of PHF1 | | Descriptor: | Histone H3.1t peptide, PHD finger protein 1, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Min, J.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-03-26 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for histone variant H3tK27me3 recognition by PHF1 and PHF19.
Elife, 9, 2020
|
|
6PI7
 
 | |
6XKC
 
 | | Crystal structure of E3 ligase | | Descriptor: | Protein fem-1 homolog C | | Authors: | Yan, X, Dong, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Min, J.R, Dong, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-26 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Molecular basis for ubiquitin ligase CRL2 FEM1C -mediated recognition of C-degron.
Nat.Chem.Biol., 17, 2021
|
|
6XCG
 
 | | Histone-lysine N-methyltransferase NSD2-PWWP1 with compound UNC6934 | | Descriptor: | Histone-lysine N-methyltransferase NSD2, N-cyclopropyl-3-oxo-N-({4-[(pyrimidin-4-yl)carbamoyl]phenyl}methyl)-3,4-dihydro-2H-1,4-benzoxazine-7-carboxamide, UNKNOWN ATOM OR ION | | Authors: | Zhou, M.Q, Dong, A, Ingerman, L.A, Hanley, R.P, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-08 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A chemical probe targeting the PWWP domain alters NSD2 nucleolar localization.
Nat.Chem.Biol., 18, 2022
|
|
8SP6
 
 | | COMPLEX STRUCTURE OF CDYL2 WITH AN ANTAGONIST | | Descriptor: | (1s,4s)-bicyclo[2.2.1]heptane, (5R0)FAL(MLZ)(5R5), Chromodomain Y-like protein 2, ... | | Authors: | Song, X, Beldar, S, Dong, A, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-05-02 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | COMPLEX STRUCTURE OF CDYL2 WITH AN ANTAGONIST
To be published
|
|
8SIG
 
 | | Crystal Structure of PRMT4 with Compound YD1-288 | | Descriptor: | 5'-{(3-aminopropyl)[2-(benzylcarbamamido)ethyl]amino}-5'-deoxyadenosine, Histone-arginine methyltransferase CARM1, SODIUM ION, ... | | Authors: | Song, X, Dong, A, Deng, Y, Huang, R, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal Structure of PRMT4 with Compound YD1-288
To be published
|
|
5KCC
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with Oxabicyclic Heptene Sulfonamide (OBHS-N) | | Descriptor: | (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-N-phenyl-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor, NCOA2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2016-06-06 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.386 Å) | | Cite: | Full antagonism of the estrogen receptor without a prototypical ligand side chain.
Nat. Chem. Biol., 13, 2017
|
|
5KCW
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with an N-trifluoroethyl OBHS-N derivative | | Descriptor: | (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-N-phenyl-N-(2,2,2-trifluoroethyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor, NCOA2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Dharmarajan, V, Goswami, D, Kastrati, I, Novick, S, Nowak, J, Zhou, H.B, Boonmuen, N, Zhao, Y, Min, J, Frasor, J, Katzenellenbogen, B.S, Griffin, P.R, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-06-07 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | Full antagonism of the estrogen receptor without a prototypical ligand side chain.
Nat. Chem. Biol., 13, 2017
|
|
5KD9
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with an N-trifluoroethyl 4-chlorobenzyl OBHS-N derivative | | Descriptor: | (1S,2R,4S)-N-(4-chlorophenyl)-5,6-bis(4-hydroxyphenyl)-N-(2,2,2-trifluoroethyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor, NCOA2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Dharmarajan, V, Goswami, D, Kastrati, I, Novick, S, Nowak, J, Zhou, H.B, Boonmuen, N, Zhao, Y, Min, J, Frasor, J, Katzenellenbogen, B.S, Griffin, P.R, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-06-07 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Full antagonism of the estrogen receptor without a prototypical ligand side chain.
Nat. Chem. Biol., 13, 2017
|
|
6WZZ
 
 | | GID4 in complex with VGLWKS peptide | | Descriptor: | Glucose-induced degradation protein 4 homolog, UNKNOWN ATOM OR ION, VGLWKS peptide | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-14 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Recognition of nonproline N-terminal residues by the Pro/N-degron pathway.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6WZX
 
 | | GID4 in complex with IGLWKS peptide | | Descriptor: | Glucose-induced degradation protein 4 homolog, ILE-GLY-LEU-TRP-LYS peptide, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-14 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Recognition of nonproline N-terminal residues by the Pro/N-degron pathway.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5KCE
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with an N-methyl, 2-chlorobenzyl OBHS-N derivative | | Descriptor: | (1S,2R,4S)-N-(2-chlorophenyl)-5,6-bis(4-hydroxyphenyl)-N-methyl-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Dharmarajan, V, Goswami, D, Kastrati, I, Novick, S, Nowak, J, Zhou, H.B, Boonmuen, N, Zhao, Y, Min, J, Frasor, J, Katzenellenbogen, B.S, Griffin, P.R, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-06-06 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.847 Å) | | Cite: | Full antagonism of the estrogen receptor without a prototypical ligand side chain.
Nat. Chem. Biol., 13, 2017
|
|
5KCU
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with an N-ethyl, alpha-naphthyl OBHS-N derivative | | Descriptor: | (1S,2R,4S)-N-ethyl-5,6-bis(4-hydroxyphenyl)-N-(naphthalen-2-yl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor, NCOA2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Dharmarajan, V, Goswami, D, Kastrati, I, Novick, S, Nowak, J, Zhou, H.B, Boonmuen, N, Zhao, Y, Min, J, Frasor, J, Katzenellenbogen, B.S, Griffin, P.R, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-06-07 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Full antagonism of the estrogen receptor without a prototypical ligand side chain.
Nat. Chem. Biol., 13, 2017
|
|
