8W1M
 
 | |
3LSJ
 
 | |
4XEK
 
 | | Pyk2-FAT domain in complex with leupaxin LD4 motif | | Descriptor: | 19-mer peptide containing Leupaxin LD4 motif, Protein-tyrosine kinase 2-beta | | Authors: | Miller, D.J. | | Deposit date: | 2014-12-24 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.793 Å) | | Cite: | Structural Basis for the Interaction between Pyk2-FAT Domain and Leupaxin LD Repeats.
Biochemistry, 55, 2016
|
|
4XEV
 
 | |
4XEF
 
 | | Pyk2-FAT complexed with Leupaxin LD motif LD1 | | Descriptor: | 20-mer peptide containing LD1 motif of leupaxin, Protein-tyrosine kinase 2-beta | | Authors: | Miller, D.J. | | Deposit date: | 2014-12-23 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for the Interaction between Pyk2-FAT Domain and Leupaxin LD Repeats.
Biochemistry, 55, 2016
|
|
7N9G
 
 | | Crystal structure of the Abl 1b Kinase domain in complex with Dasatinib and Imatinib | | Descriptor: | 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, N-(2-CHLORO-6-METHYLPHENYL)-2-({6-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]-2-METHYLPYRIMIDIN-4-YL}AMINO)-1,3-THIAZOLE-5-CARBOXAMIDE, PHOSPHATE ION, ... | | Authors: | Miller, D.J, Xie, T. | | Deposit date: | 2021-06-17 | | Release date: | 2022-04-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Imatinib can act as an Allosteric Activator of Abl Kinase.
J.Mol.Biol., 434, 2022
|
|
2QVL
 
 | |
8G66
 
 | | Structure with SJ3149 | | Descriptor: | (3S)-3-{5-[(1,2-benzoxazol-3-yl)amino]-1-oxo-1,3-dihydro-2H-isoindol-2-yl}piperidine-2,6-dione, Casein kinase I isoform alpha, DNA damage-binding protein 1, ... | | Authors: | Miller, D.J, Young, S.M, Fischer, M. | | Deposit date: | 2023-02-14 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Structure of ternary complex with molecular glue targeting CK1A for degradation by the CRL4CRBN ubiquitin ligase
To Be Published
|
|
6WJH
 
 | |
3R15
 
 | | Structure Treponema Denticola Factor H Binding Protein | | Descriptor: | Factor H binding protein, THIOCYANATE ION | | Authors: | Miller, D.P, McDowell, J.V, Burgner, J, Heroux, A, Bell, J.K, Marconi, R.T. | | Deposit date: | 2011-03-09 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Structure Treponema Denticola Factor H Binding Protein
To be Published
|
|
2QV7
 
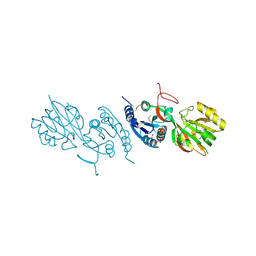 | | Crystal Structure of Diacylglycerol Kinase DgkB in complex with ADP and Mg | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Diacylglycerol Kinase DgkB, MAGNESIUM ION | | Authors: | Miller, D.J, Jerga, A, Rock, C.O, White, S.W. | | Deposit date: | 2007-08-07 | | Release date: | 2008-06-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Analysis of the Staphylococcus aureus DgkB Structure Reveals a Common Catalytic Mechanism for the Soluble Diacylglycerol Kinases.
Structure, 16, 2008
|
|
5W93
 
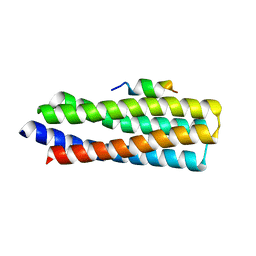 | | p130Cas complex with paxillin LD1 | | Descriptor: | Breast cancer anti-estrogen resistance protein 1, Paxillin | | Authors: | Miller, D.J, Zheng, J.J. | | Deposit date: | 2017-06-22 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural and functional insights into the interaction between the Cas family scaffolding protein p130Cas and the focal adhesion-associated protein paxillin.
J. Biol. Chem., 292, 2017
|
|
3QZ0
 
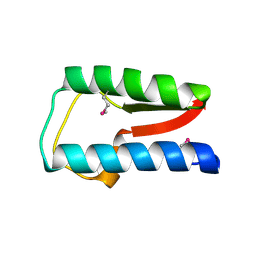 | | Structure of Treponema denticola Factor H Binding protein (FhbB), selenomethionine derivative | | Descriptor: | Factor H binding protein, GLYCEROL, THIOCYANATE ION | | Authors: | Miller, D.P, McDowell, J.V, Heroux, A, Bell, J.K, Marconi, R.T, Conrad, D.H, Burgner, J.W. | | Deposit date: | 2011-03-04 | | Release date: | 2012-03-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure of factor H-binding protein B (FhbB) of the periopathogen, Treponema denticola: insights into progression of periodontal disease.
J.Biol.Chem., 287, 2012
|
|
6ASR
 
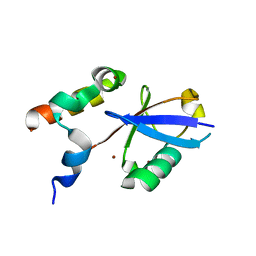 | | REV1 UBM2 domain complex with ubiquitin | | Descriptor: | DNA repair protein REV1, NICKEL (II) ION, Ubiquitin | | Authors: | Miller, D.J. | | Deposit date: | 2017-08-25 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.356 Å) | | Cite: | Structures of REV1 UBM2 Domain Complex with Ubiquitin and with a Small-Molecule that Inhibits the REV1 UBM2-Ubiquitin Interaction.
J. Mol. Biol., 430, 2018
|
|
2GQD
 
 | |
5UDH
 
 | | HHARI/ARIH1-UBCH7~Ubiquitin | | Descriptor: | E3 ubiquitin-protein ligase ARIH1, Ubiquitin C variant, Ubiquitin-conjugating enzyme E2 L3, ... | | Authors: | Miller, D.J, Schulman, B.A. | | Deposit date: | 2016-12-27 | | Release date: | 2017-06-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Structural Studies of HHARI/UbcH7Ub Reveal Unique E2Ub Conformational Restriction by RBR RING1.
Structure, 25, 2017
|
|
3LSR
 
 | |
1Q7R
 
 | |
1TT4
 
 | | Structure of NP459575, a predicted glutathione synthase from Salmonella typhimurium | | Descriptor: | MAGNESIUM ION, SULFATE ION, putative cytoplasmic protein | | Authors: | Miller, D.J, Shuvalova, L, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-06-21 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structure of NP459575, a predicted glutathione synthase from Salmonella typhimurium
To be Published
|
|
6NJ7
 
 | | 11-BETA DEHYDROGENASE ISOZYME 1 IN COMPLEX WITH COLLETOIC ACID | | Descriptor: | (1S,4S,5S,9S)-9-hydroxy-8-methyl-4-(propan-2-yl)spiro[4.5]dec-7-ene-1-carboxylic acid, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Miller, D.J, Rivas, F. | | Deposit date: | 2019-01-02 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mechanistic Insight on the Mode of Action of Colletoic Acid.
J.Med.Chem., 62, 2019
|
|
1N2X
 
 | | Crystal Structure Analysis of TM0872, a Putative SAM-dependent Methyltransferase, Complexed with SAM | | Descriptor: | S-ADENOSYLMETHIONINE, S-adenosyl-methyltransferase mraW, SULFATE ION | | Authors: | Miller, D.J, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-10-24 | | Release date: | 2003-01-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal complexes of a predicted S-adenosylmethionine-dependent methyltransferase reveal a typical AdoMet binding domain and a substrate recognition domain
Protein Sci., 12, 2003
|
|
6NXK
 
 | | Ubiquitin binding variants | | Descriptor: | Anaphase-promoting complex subunit 2, Polyubiquitin-C | | Authors: | Miller, D.J, Watson, E.R. | | Deposit date: | 2019-02-08 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Protein engineering of a ubiquitin-variant inhibitor of APC/C identifies a cryptic K48 ubiquitin chain binding site.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6NXL
 
 | | Ubiquitin binding variants | | Descriptor: | Polyubiquitin-B | | Authors: | Miller, D.J, Watson, E.R. | | Deposit date: | 2019-02-08 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Protein engineering of a ubiquitin-variant inhibitor of APC/C identifies a cryptic K48 ubiquitin chain binding site.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
2B4Q
 
 | | Pseudomonas aeruginosa RhlG/NADP active-site complex | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Rhamnolipids biosynthesis 3-oxoacyl-[acyl-carrier-protein] reductase | | Authors: | Miller, D.J, White, S.W. | | Deposit date: | 2005-09-26 | | Release date: | 2006-05-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of RhlG, an Essential beta-Ketoacyl Reductase in the Rhamnolipid Biosynthetic Pathway of Pseudomonas aeruginosa.
J.Biol.Chem., 281, 2006
|
|
1SU1
 
 | | Structural and biochemical characterization of Yfce, a phosphoesterase from E. coli | | Descriptor: | Hypothetical protein yfcE, SULFATE ION, ZINC ION | | Authors: | Miller, D.J, Shuvalova, L, Evdokimova, E, Savchenko, A, Yakunin, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-03-25 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and biochemical characterization of a novel Mn2+-dependent phosphodiesterase encoded by the yfcE gene.
Protein Sci., 16, 2007
|
|
