5ES2
 
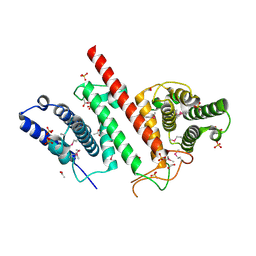 | | The crystal structure of a functionally uncharacterized protein LPG0634 from Legionella pneumophila subsp. pneumophila str. Philadelphia 1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, SULFATE ION, ... | | Authors: | Tan, K, Xu, X, Cui, H, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-11-16 | | Release date: | 2015-12-16 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of a functionally uncharacterized protein LPG0634 from Legionella pneumophila subsp. pneumophila str. Philadelphia 1
To Be Published
|
|
6UAM
 
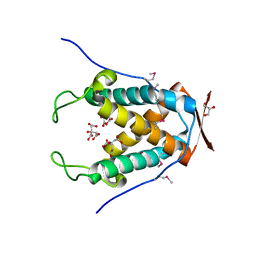 | |
6UG5
 
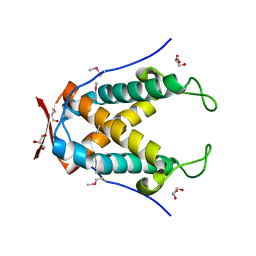 | |
4S1P
 
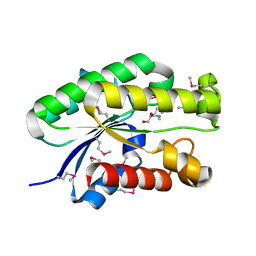 | | Shel_16390 protein, a putative SGNH hydrolase from Slackia heliotrinireducens | | Descriptor: | UNKNOWN LIGAND, Uncharacterized protein | | Authors: | Osipiuk, J, Cuff, M.E, Wu, R, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-01-14 | | Release date: | 2015-01-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Shel_16390 protein, a putative SGNH hydrolase from Slackia heliotrinireducens
To be Published
|
|
4RYK
 
 | | Crystal structure of a putative transcriptional regulator from Listeria monocytogenes EGD-e | | Descriptor: | DI(HYDROXYETHYL)ETHER, L(+)-TARTARIC ACID, Lmo0325 protein, ... | | Authors: | Filippova, E.V, Wawrzak, Z, Minasov, G, Kiryukhina, O, Jedrzejczak, R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-12-15 | | Release date: | 2015-01-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of a putative transcriptional regulator from Listeria monocytogenes EGD-e
To be Published
|
|
5CQF
 
 | | Crystal structure of L-lysine 6-monooxygenase from Pseudomonas syringae | | Descriptor: | IODIDE ION, L-lysine 6-monooxygenase | | Authors: | Michalska, K, Bigelow, L, Jedrzejczak, R, Weerth, R.S, Cao, H, Yennamalli, R, Phillips Jr, G.N, Thomas, M.G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-07-21 | | Release date: | 2015-09-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of L-lysine 6-monooxygenase from Pseudomonas syringae
To Be Published
|
|
5DU2
 
 | | Structural analysis of EspG2 glycosyltransferase | | Descriptor: | EspG2 glycosyltransferase | | Authors: | Michalska, K, Elshahawi, S.I, Bigelow, L, Babnigg, G, Thorson, J.S, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-09-18 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural analysis of EspG2 glycosyltransferase
To Be Published
|
|
5DS0
 
 | | Crystal structure of TET aminopeptidase from marine sediment archaeon Thaumarchaeota archaeon SCGC AB-539-E09 | | Descriptor: | COBALT (II) ION, GLYCEROL, Peptidase M42 | | Authors: | Michalska, K, Chhor, G, Mootz, J, Endres, M, Jedrzejczak, R, Babnigg, G, Steen, A, Lloyd, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-09-16 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of TET aminopeptidase from marine sediment archaeon Thaumarchaeota archaeon SCGC AB-539-E09
To Be Published
|
|
5I4Q
 
 | | Contact-dependent inhibition system from Escherichia coli NC101 - ternary CdiA/CdiI/EF-Tu complex (domains 2 and 3) | | Descriptor: | CHLORIDE ION, Contact-dependent inhibitor A, Contact-dependent inhibitor I, ... | | Authors: | Michalska, K, Stols, L, Eschenfeldt, W, Hayes, C.S, Goulding, C.W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI) | | Deposit date: | 2016-02-12 | | Release date: | 2017-06-28 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of a novel antibacterial toxin that exploits elongation factor Tu to cleave specific transfer RNAs.
Nucleic Acids Res., 45, 2017
|
|
5DA8
 
 | | Crystal structure of chaperonin GroEL from | | Descriptor: | 60 kDa chaperonin, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Chang, C, Marshall, N, Feldmann, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-08-19 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of chaperonin GroEL from
To Be Published
|
|
5EVI
 
 | | Crystal Structure of Beta-Lactamase/D-Alanine Carboxypeptidase from Pseudomonas syringae | | Descriptor: | 1,2-ETHANEDIOL, Beta-Lactamase/D-Alanine Carboxypeptidase, SULFATE ION | | Authors: | Kim, Y, Hatzos-Skintges, C, Endres, M, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-11-19 | | Release date: | 2016-01-13 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Beta-Lactamase/D-Alanine Carboxypeptidase from Pseudomonas syringae
To Be Published
|
|
5I4R
 
 | | Contact-dependent inhibition system from Escherichia coli NC101 - ternary CdiA/CdiI/EF-Tu complex (trypsin-modified) | | Descriptor: | Contact-dependent inhibitor A, Contact-dependent inhibitor I, Elongation factor Tu, ... | | Authors: | Michalska, K, Stols, L, Eschenfeldt, W, Hayes, C.S, Goulding, C.W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI) | | Deposit date: | 2016-02-12 | | Release date: | 2017-06-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of a novel antibacterial toxin that exploits elongation factor Tu to cleave specific transfer RNAs.
Nucleic Acids Res., 45, 2017
|
|
6UHE
 
 | |
5C4Y
 
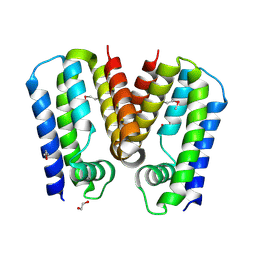 | | Crystal structure of putative TetR family transcription factor from Listeria monocytogenes | | Descriptor: | 1,2-ETHANEDIOL, Putative transcription regulator Lmo0852 | | Authors: | Chang, C, Tesar, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-06-18 | | Release date: | 2015-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of putative TetR family transcription factor from Listeria monocytogenes
to be published
|
|
6UHI
 
 | |
5C6U
 
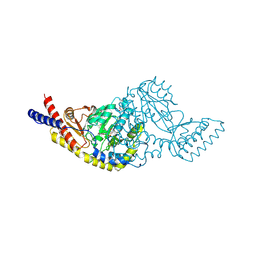 | | Rv3722c aminotransferase from Mycobacterium tuberculosis | | Descriptor: | Aminotransferase, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | OSIPIUK, J, Hatzos-Skintges, C, Jedrzejczak, R, Babnigg, G, Sacchettini, J, JOACHIMIAK, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2015-06-23 | | Release date: | 2015-07-15 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Rv3722c aminotransferase from Mycobacterium tuberculosis.
to be published
|
|
5E2E
 
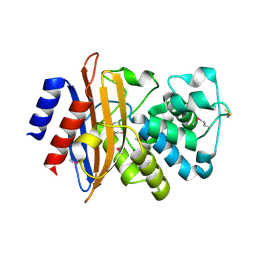 | | Crystal Structure of Beta-lactamase Precursor BlaA from Yersinia enterocolitica | | Descriptor: | Beta-lactamase | | Authors: | Kim, Y, Joachimiak, G, Endres, M, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-10-01 | | Release date: | 2015-10-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Beta-lactamase Precursor BlaA from Yersinia enterocolitica
To Be Published
|
|
5E2C
 
 | | Crystal structure of N-terminal domain of cytoplasmic peptidase PepQ from Mycobacterium tuberculosis H37Rv | | Descriptor: | Xaa-Pro dipeptidase | | Authors: | Chang, C, Endres, L, Endres, M, SACCHETTINI, J, JOACHIMIAK, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2015-09-30 | | Release date: | 2015-10-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of N-terminal domain of cytoplasmic peptidase PepQ from Mycobacterium tuberculosis H37Rv
To Be Published
|
|
5E2G
 
 | | Crystal Structure of D-alanine Carboxypeptidase AmpC from Burkholderia cenocepacia | | Descriptor: | ACETIC ACID, Beta-lactamase, THIOCYANATE ION | | Authors: | Kim, Y, Joachimiak, G, Endres, M, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-10-01 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Crystal Structure of D-alanine Carboxypeptidase AmpC from Burkholderia cenocepacia
To Be Published
|
|
5ER3
 
 | | Crystal structure of ABC transporter system solute-binding protein from Rhodopirellula baltica SH 1 | | Descriptor: | CALCIUM ION, GLYCEROL, Sugar ABC transporter, ... | | Authors: | Chang, C, Duke, N, Endres, M, Mack, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-11-13 | | Release date: | 2015-11-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | Crystal structure of ABC transporter system solute-binding protein from Rhodopirellula baltica SH 1
To Be Published
|
|
5DUK
 
 | |
5DYF
 
 | | The crystal structure of Aminopeptidase N in complex with N-benzyl-1,2-diaminoethylphosphonic acid | | Descriptor: | Aminopeptidase N, GLYCEROL, IMIDAZOLE, ... | | Authors: | Nocek, B, Joachimiak, A, Vassiliou, S, Berlicki, L, Mucha, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-09-24 | | Release date: | 2015-11-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.854 Å) | | Cite: | The crystal structure of Aminopeptidase N in complex with N-benzyl-1,2-diaminoethylphosphonic acid
To Be Published
|
|
5EVH
 
 | | Crystal structure of known function protein from Kribbella flavida DSM 17836 | | Descriptor: | GLYCEROL, Uncharacterized protein | | Authors: | Chang, C, Duke, N, Endres, M, Chhor, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-11-19 | | Release date: | 2015-12-02 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | Crystal structure of known function protein from Kribbella flavida DSM 17836
To Be Published
|
|
5F2H
 
 | | 2.75 Angstrom resolution crystal structure of uncharacterized protein from Bacillus cereus ATCC 10987 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Uncharacterized protein | | Authors: | Halavaty, A.S, Filippova, E.V, Wawrzak, Z, Minasov, G, Kiryukhina, O, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-12-01 | | Release date: | 2015-12-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | 2.75 Angstrom resolution crystal structure of uncharacterized protein from Bacillus cereus ATCC 10987
To Be Published
|
|
6AZY
 
 | | Crystal structure of Hsp104 R328M/R757M mutant from Calcarisporiella thermophila | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Heat shock protein Hsp104 | | Authors: | Michalska, K, Bigelow, L, Hatzos-Skintges, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-09-13 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of Calcarisporiella thermophila Hsp104 Disaggregase that Antagonizes Diverse Proteotoxic Misfolding Events.
Structure, 27, 2019
|
|
