6Q40
 
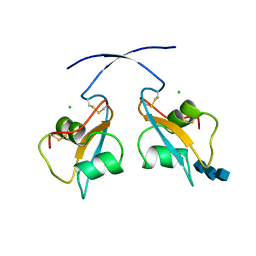 | | A secreted LysM effector of the wheat pathogen Zymoseptoria tritici protects the fungal hyphae against chitinase hydrolysis through ligand-dependent polymerisation of LysM homodimers | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, LysM domain-containing protein | | Authors: | Mesters, J.R, Saleem-Batcha, R, Sanchez-Vallet, A, Thomma, B.P.H.J. | | Deposit date: | 2018-12-05 | | Release date: | 2019-10-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.412 Å) | | Cite: | A secreted LysM effector protects fungal hyphae through chitin-dependent homodimer polymerization.
Plos Pathog., 16, 2020
|
|
2JBK
 
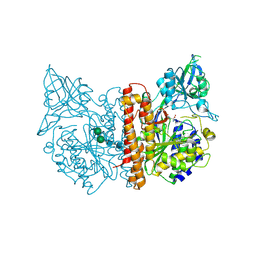 | | membrane-bound glutamate carboxypeptidase II (GCPII) in complex with quisqualic acid (quisqualate, alpha-amino-3,5-dioxo-1,2,4- oxadiazolidine-2-propanoic acid) | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mesters, J.R, Henning, K, Hilgenfeld, R. | | Deposit date: | 2006-12-07 | | Release date: | 2006-12-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Human Glutamate Carboxypeptidase II Inhibition: Structures of Gcpii in Complex with Two Potent Inhibitors, Quisqualate and 2-Pmpa.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2JBJ
 
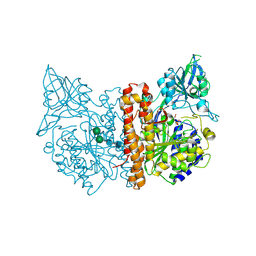 | | membrane-bound glutamate carboxypeptidase II (GCPII) in complex with 2-PMPA (2-phosphonoMethyl-pentanedioic acid) | | Descriptor: | (2S)-2-(PHOSPHONOMETHYL)PENTANEDIOIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mesters, J.R, Henning, K, Hilgenfeld, R. | | Deposit date: | 2006-12-07 | | Release date: | 2006-12-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Human Glutamate Carboxypeptidase II Inhibition: Structures of Gcpii in Complex with Two Potent Inhibitors, Quisqualate and 2-Pmpa.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2C6G
 
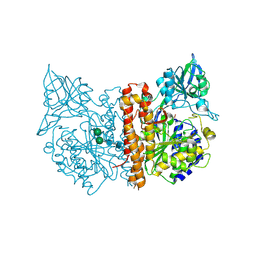 | | Membrane-bound glutamate carboxypeptidase II (GCPII) with bound glutamate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Mesters, J.R, Barinka, C, Li, W, Tsukamoto, T, Majer, P, Slusher, B.S, Konvalinka, J, Hilgenfeld, R. | | Deposit date: | 2005-11-09 | | Release date: | 2006-02-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of glutamate carboxypeptidase II, a drug target in neuronal damage and prostate cancer.
EMBO J., 25, 2006
|
|
2C6C
 
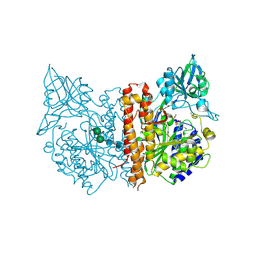 | | membrane-bound glutamate carboxypeptidase II (GCPII) in complex with GPI-18431 (S)-2-(4-iodobenzylphosphonomethyl)-pentanedioic acid | | Descriptor: | (2S)-2-{[HYDROXY(4-IODOBENZYL)PHOSPHORYL]METHYL}PENTANEDIOIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mesters, J.R, Barinka, C, Li, W, Tsukamoto, T, Majer, P, Slusher, B.S, Konvalinka, J, Hilgenfeld, R. | | Deposit date: | 2005-11-09 | | Release date: | 2006-02-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Glutamate Carboxypeptidase II, a Drug Target in Neuronal Damage and Prostate Cancer.
Embo J., 25, 2006
|
|
2C6P
 
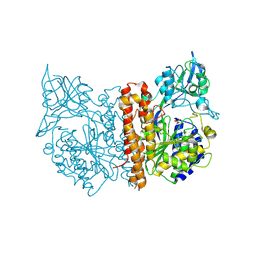 | | Membrane-bound glutamate carboxypeptidase II (GCPII) in complex with phosphate anion | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Mesters, J.R, Barinka, C, Li, W, Tsukamoto, T, Majer, P, Slusher, B.S, Konvalinka, J, Hilgenfeld, R. | | Deposit date: | 2005-11-11 | | Release date: | 2006-02-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure of Glutamate Carboxypeptidase II, a Drug Target in Neuronal Damage and Prostate Cancer.
Embo J., 25, 2006
|
|
6H2J
 
 | |
6IAH
 
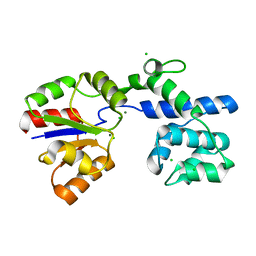 | | Phosphatase Tt82 from Thermococcus thioreducens | | Descriptor: | CHLORIDE ION, Hydrolase, MAGNESIUM ION | | Authors: | Havlickova, P, Brinsa, V, Brynda, J, Pachl, P, Prudnikova, T, Mesters, J.R, Kascakova, B, Kuty, M, Pusey, M.L, Ng, J.D, Rezacova, P, Smatanova, I.K. | | Deposit date: | 2018-11-26 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A novel structurally characterized haloacid dehalogenase superfamily phosphatase from Thermococcus thioreducens with diverse substrate specificity.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
4ZM6
 
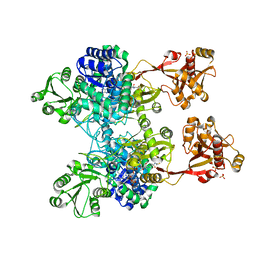 | | A unique GCN5-related glucosamine N-acetyltransferase region exist in the fungal multi-domain GH3 beta-N-acetylglucosaminidase | | Descriptor: | ACETYL COENZYME *A, N-acetyl-beta-D glucosaminidase, SULFATE ION | | Authors: | Qin, Z, Xiao, Y, Yang, X, Jiang, Z, Yang, S, Mesters, J.R. | | Deposit date: | 2015-05-02 | | Release date: | 2015-12-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A unique GCN5-related glucosamine N-acetyltransferase region exist in the fungal multi-domain glycoside hydrolase family 3 beta-N-acetylglucosaminidase
Sci Rep, 5, 2015
|
|
1HA3
 
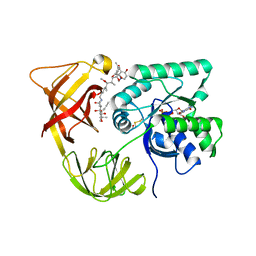 | | ELONGATION FACTOR TU IN COMPLEX WITH aurodox | | Descriptor: | BETA-MERCAPTOETHANOL, ELONGATION FACTOR TU, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Vogeley, L, Palm, G.J, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2001-03-26 | | Release date: | 2001-05-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational Change of Elongation Factor TU Induced by Antibiotic Binding: Crystal Structure of the Complex between EF-TU:Gdp and Aurodox
J.Biol.Chem., 276, 2001
|
|
4P16
 
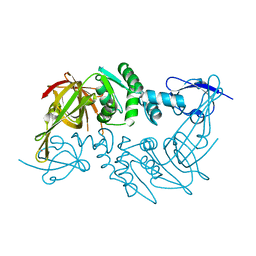 | | Crystal structure of the papain-like protease of Middle-East Respiratory Syndrome coronavirus | | Descriptor: | ORF1a, ZINC ION | | Authors: | Lei, J, Mesters, J.R, Ma, Q, Hilgenfeld, R. | | Deposit date: | 2014-02-25 | | Release date: | 2014-05-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the papain-like protease of MERS coronavirus reveals unusual, potentially druggable active-site features.
Antiviral Res., 109C, 2014
|
|
3RK4
 
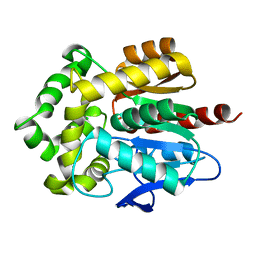 | | Structure of Rhodococcus rhodochrous haloalkane dehalogenase mutant DhaA31 | | Descriptor: | CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Lahoda, M, Stsiapanava, A, Mesters, J, Chaloupkova, R, Damborsky, J, Kuta Smatanova, I. | | Deposit date: | 2011-04-17 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Crystallographic analysis of 1,2,3-trichloropropane biodegradation by the haloalkane dehalogenase DhaA31.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4FWB
 
 | | Structure of Rhodococcus rhodochrous haloalkane dehalogenase mutant DhaA31 in complex with 1, 2, 3 - trichloropropane | | Descriptor: | 1,2,3-trichloropropane, CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Lahoda, M, Stsiapanava, A, Mesters, J, Kuta Smatanova, I. | | Deposit date: | 2012-06-30 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Crystallographic analysis of 1,2,3-trichloropropane biodegradation by the haloalkane dehalogenase DhaA31.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4B9H
 
 | | Cladosporium fulvum LysM effector Ecp6 in complex with a beta-1,4- linked N-acetyl-D-glucosamine tetramer: I3C heavy atom derivative | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, ... | | Authors: | Saleem-Batcha, R, Sanchez-Vallet, A, Hansen, G, Thomma, B.P.H.J, Mesters, J.R. | | Deposit date: | 2012-09-04 | | Release date: | 2013-07-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Fungal Effector Ecp6 Outcompetes Host Immune Receptor for Chitin Binding Through Intrachain Lysm Dimerization
Elife, 2, 2013
|
|
2XCU
 
 | | Membrane-embedded monofunctional glycosyltransferase WaaA of Aquifex aeolicus, complex with CMP | | Descriptor: | 3-DEOXY-D-MANNO-2-OCTULOSONIC ACID TRANSFERASE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Schmidt, H, Hansen, G, Hilgenfeld, R, Mamat, U, Mesters, J.R. | | Deposit date: | 2010-04-26 | | Release date: | 2011-05-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural and Mechanistic Analysis of the Membrane-Embedded Glycosyltransferase Waaa Required for Lipopolysaccharide Synthesis.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4B8V
 
 | | Cladosporium fulvum LysM effector Ecp6 in complex with a beta-1,4- linked N-acetyl-D-glucosamine tetramer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, ... | | Authors: | Saleem-Batcha, R, Sanchez-Vallet, A, Hansen, G, Thomma, B.P.H.J, Mesters, J.R. | | Deposit date: | 2012-08-30 | | Release date: | 2013-07-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Fungal Effector Ecp6 Outcompetes Host Immune Receptor for Chitin Binding Through Intrachain Lysm Dimerization
Elife, 2, 2013
|
|
2XCI
 
 | | Membrane-embedded monofunctional glycosyltransferase WaaA of Aquifex aeolicus, substrate-free form | | Descriptor: | 3-DEOXY-D-MANNO-2-OCTULOSONIC ACID TRANSFERASE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Schmidt, H, Hansen, G, Hilgenfeld, R, Mamat, U, Mesters, J.R. | | Deposit date: | 2010-04-26 | | Release date: | 2011-05-11 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Mechanistic Analysis of the Membrane-Embedded Glycosyltransferase Waaa Required for Lipopolysaccharide Synthesis.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1P9U
 
 | | Coronavirus Main Proteinase (3CLpro) Structure: Basis for Design of anti-SARS Drugs | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, PHQ-VNSTLQ-CHLOROMETHYLKETONE INHIBITOR, SULFATE ION, ... | | Authors: | Anand, K, Ziebuhr, J, Wadhwani, P, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2003-05-12 | | Release date: | 2003-05-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Coronavirus Main Proteinase (3CLpro) Structure: Basis for Design of anti-SARS Drugs
Science, 300, 2003
|
|
1P9S
 
 | | Coronavirus Main Proteinase (3CLpro) Structure: Basis for Design of anti-SARS Drugs | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Replicase polyprotein 1ab | | Authors: | Anand, K, Ziebuhr, J, Wadhwani, P, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2003-05-12 | | Release date: | 2003-05-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Coronavirus Main Proteinase (3CLpro) Structure: Basis for Design of anti-SARS Drugs
Science, 300, 2003
|
|
3RK6
 
 | |
1EXM
 
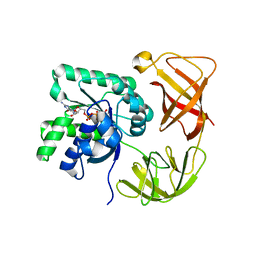 | | CRYSTAL STRUCTURE OF THERMUS THERMOPHILUS ELONGATION FACTOR TU (EF-TU) IN COMPLEX WITH THE GTP ANALOGUE GPPNHP. | | Descriptor: | ELONGATION FACTOR TU (EF-TU), MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Hogg, T, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2000-05-03 | | Release date: | 2000-06-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Insights into the GTPase Mechanism of EF-Tu from Structural Studies
The Ribosome: Structure, Function, Antibiotics, and Cellular Interactions, 28, 2000
|
|
4HZG
 
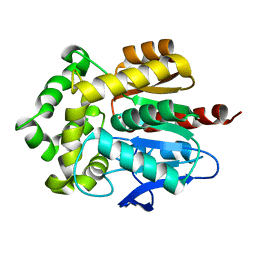 | | Structure of haloalkane dehalogenase DhaA from Rhodococcus rhodochrous | | Descriptor: | CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Stsiapanava, A, Weiss, M.S, Mesters, J.R, Kuta Smatanova, I. | | Deposit date: | 2012-11-15 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystallographic analysis of 1,2,3-trichloropropane biodegradation by the haloalkane dehalogenase DhaA31.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1IZH
 
 | | Inhibitor of HIV protease with unusual binding mode potently inhibiting multi-resistant protease mutants | | Descriptor: | proteinase, {(1S)-1-BENZYL-4-[3-CARBAMOYL-1-(1-CARBAMOYL-2-PHENYL-ETHYLCARBAMOYL)-(S)-PROPYLCARBAMOYL]-2-OXO-5-PHENYL-PENTYL}-CARBAMIC ACID TERT-BUTYL ESTER | | Authors: | Weber, J, Mesters, J.R, Lepsik, M, Prejdova, J, Svec, M, Sponarova, J, Mlcochova, P, Skalicka, K, Strisovsky, K, Uhlikova, T, Soucek, M, Machala, L, Stankova, M, Vondrasek, J, Klimkait, T, Kraeusslich, H.-G, Hilgenfeld, R, Konvalinka, J. | | Deposit date: | 2002-10-02 | | Release date: | 2002-12-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unusual Binding Mode of an HIV-1 Protease Inhibitor Explains its Potency against Multi-drug-resistant Virus Strains
J.MOL.BIOL., 324, 2002
|
|
1IZI
 
 | | Inhibitor of HIV protease with unusual binding mode potently inhibiting multi-resistant protease mutants | | Descriptor: | CHLORIDE ION, proteinase, {(1S)-1-BENZYL-4-[3-CARBAMOYL-1-(1-CARBAMOYL-2-PHENYL-ETHYLCARBAMOYL)-(S)-PROPYLCARBAMOYL]-2-OXO-5-PHENYL-PENTYL}-CARBAMIC ACID TERT-BUTYL ESTER | | Authors: | Weber, J, Mesters, J.R, Lepsik, M, Prejdova, J, Svec, M, Sponarova, J, Mlcochova, P, Skalicka, K, Strisovsky, K, Uhlikova, T, Soucek, M, Machala, L, Stankova, M, Vondrasek, J, Klimkait, T, Kraeusslich, H.-G, Hilgenfeld, R, Konvalinka, J. | | Deposit date: | 2002-10-02 | | Release date: | 2002-12-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Unusual Binding Mode of an HIV-1 Protease Inhibitor Explains its Potency against Multi-drug-resistant Virus Strains
J.MOL.BIOL., 324, 2002
|
|
1LVO
 
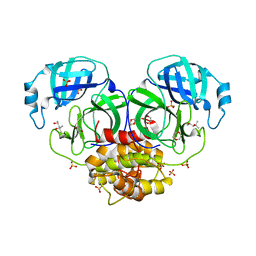 | | Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra alpha-helical domain | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1,4-DIETHYLENE DIOXIDE, Replicase, ... | | Authors: | Anand, K, Palm, G.J, Mesters, J.R, Siddell, S.G, Ziebuhr, J, Hilgenfeld, R. | | Deposit date: | 2002-05-29 | | Release date: | 2002-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra alpha-helical domain.
EMBO J., 21, 2002
|
|
