7Z38
 
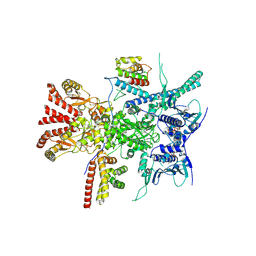 | | Structure of the RAF1-HSP90-CDC37 complex (RHC-I) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein HSP 90-beta, Hsp90 co-chaperone Cdc37, ... | | Authors: | Mesa, P, Garcia-Alonso, S, Barbacid, M, Montoya, G. | | Deposit date: | 2022-03-01 | | Release date: | 2022-09-14 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structure of the RAF1-HSP90-CDC37 complex reveals the basis of RAF1 regulation.
Mol.Cell, 82, 2022
|
|
7Z37
 
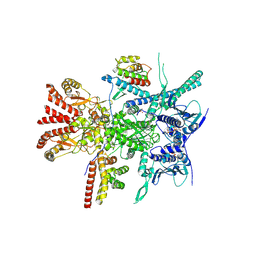 | | Structure of the RAF1-HSP90-CDC37 complex (RHC-II) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein HSP 90-beta, Hsp90 co-chaperone Cdc37, ... | | Authors: | Mesa, P, Garcia-Alonso, S, Barbacid, M, Montoya, G. | | Deposit date: | 2022-03-01 | | Release date: | 2022-09-14 | | Last modified: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Structure of the RAF1-HSP90-CDC37 complex reveals the basis of RAF1 regulation.
Mol.Cell, 82, 2022
|
|
6GTC
 
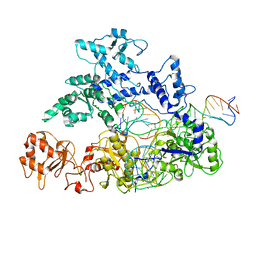 | | Transition state structure of Cpf1(Cas12a) I1 conformation | | Descriptor: | CRISPR-associated endonuclease Cas12a, DNA (5'-D(P*CP*GP*AP*GP*CP*TP*CP*GP*TP*TP*AP*GP*AP*GP*AP*AP*G)-3'), DNA (5'-D(P*TP*GP*AP*CP*TP*TP*CP*TP*CP*TP*AP*AP*CP*AP*AP*GP*CP*TP*CP*G)-3'), ... | | Authors: | Mesa, P, Montoya, G. | | Deposit date: | 2018-06-18 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Conformational Activation Promotes CRISPR-Cas12a Catalysis and Resetting of the Endonuclease Activity.
Cell, 175, 2018
|
|
6GTG
 
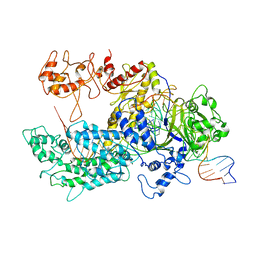 | | Transition state structure of Cpf1(Cas12a) I4 conformation | | Descriptor: | CRISPR-associated endonuclease Cas12a, DNA (32-MER), DNA (5'-D(P*CP*GP*AP*GP*CP*TP*CP*GP*TP*TP*AP*GP*AP*GP*AP*AP*GP*T)-3'), ... | | Authors: | Mesa, P, Montoya, G, Stella, S. | | Deposit date: | 2018-06-18 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Conformational Activation Promotes CRISPR-Cas12a Catalysis and Resetting of the Endonuclease Activity.
Cell, 175, 2018
|
|
5JNO
 
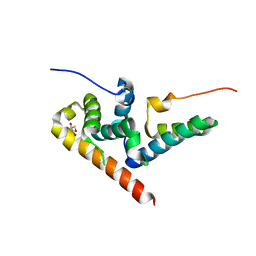 | |
1NO1
 
 | | Structure of truncated variant of B.subtilis SPP1 phage G39P helicase loader/inhibitor protein | | Descriptor: | replisome organizer | | Authors: | Bailey, S, Sedelnikova, S.E, Mesa, P, Ayora, S, Waltho, J.P, Ashcroft, A.E, Baron, A.J, Alonso, J.C, Rafferty, J.B. | | Deposit date: | 2003-01-15 | | Release date: | 2003-05-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural analysis of Bacillus subtilis SPP1 phage helicase loader protein G39P
J.Biol.Chem., 278, 2003
|
|
8AXA
 
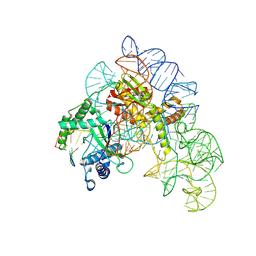 | | Cryo-EM structure of shCas12k-sgRNA-dsDNA ternary complex (type V-K CRISPR-associated transposon) | | Descriptor: | Cas12k, DNA non-target strand, DNA target strand, ... | | Authors: | Tenjo-Castano, F, Sofos, N, Stella, S, Fuglsang, A, Pape, T, Mesa, P, Stutzke, L.S, Temperini, P, Montoya, G. | | Deposit date: | 2022-08-31 | | Release date: | 2024-04-10 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Conformational landscape of the type V-K CRISPR-associated transposon integration assembly.
Mol.Cell, 84, 2024
|
|
2XSM
 
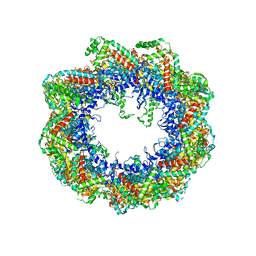 | | Crystal structure of the mammalian cytosolic chaperonin CCT in complex with tubulin | | Descriptor: | CCT | | Authors: | Munoz, I.G, Yebenes, H, Zhou, M, Mesa, P, Serna, M, Bragado-Nilsson, E, Beloso, A, Robinson, C.V, Valpuesta, J.M, Montoya, G. | | Deposit date: | 2010-10-29 | | Release date: | 2010-12-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Crystal Structure of the Open Conformation of the Mammalian Chaperonin Cct in Complex with Tubulin.
Nat.Struct.Mol.Biol., 18, 2011
|
|
8RDU
 
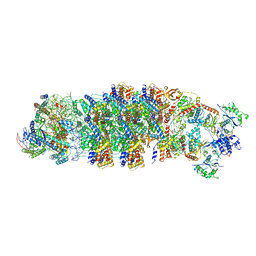 | | Conformational Landscape of the Type V-K CRISPR-associated TransposonIntegration Assembly CAST V-K composite map | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, LE, MAGNESIUM ION, ... | | Authors: | Tenjo-Castano, F, Mesa, P, Montoya, G. | | Deposit date: | 2023-12-08 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Conformational landscape of the type V-K CRISPR-associated transposon integration assembly.
Mol.Cell, 84, 2024
|
|
8RKU
 
 | | Conformational Landscape of the Type V-K CRISPR-associated TransposonIntegration Assembly CAST V-K TnsC domain local-refinement map | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Non-target strand - LE, ... | | Authors: | Tenjo-Castano, F, Mesa, P, Montoya, G. | | Deposit date: | 2023-12-30 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Conformational landscape of the type V-K CRISPR-associated transposon integration assembly.
Mol.Cell, 84, 2024
|
|
8RKV
 
 | | Conformational Landscape of the Type V-K CRISPR-associated TransposonIntegration Assembly CAST V-K TnsB domain local-refinement map | | Descriptor: | LE, MAGNESIUM ION, Non-target strand - LE, ... | | Authors: | Tenjo-Castano, F, Mesa, P, Montoya, G. | | Deposit date: | 2023-12-30 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Conformational landscape of the type V-K CRISPR-associated transposon integration assembly.
Mol.Cell, 84, 2024
|
|
8RKT
 
 | | Conformational Landscape of the Type V-K CRISPR-associated TransposonIntegration Assembly CAST V-K Cas12k domain local-refinement map | | Descriptor: | MAGNESIUM ION, Non-target strand - LE, ShCas12k, ... | | Authors: | Tenjo-Castano, F, Mesa, P, Montoya, G. | | Deposit date: | 2023-12-30 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | Conformational landscape of the type V-K CRISPR-associated transposon integration assembly.
Mol.Cell, 84, 2024
|
|
6GTE
 
 | | Transient state structure of CRISPR-Cpf1 (Cas12a) I3 conformation | | Descriptor: | CRISPR-associated endonuclease Cas12a, DNA (5'-D(P*AP*TP*GP*AP*CP*TP*TP*CP*TP*CP*TP*AP*AP*CP*AP*AP*GP*CP*TP*CP*G)-3'), DNA (5'-D(P*CP*GP*AP*GP*CP*TP*CP*GP*TP*TP*AP*GP*AP*GP*AP*AP*G)-3'), ... | | Authors: | Montoya, G, Mesa, P, Stella, S. | | Deposit date: | 2018-06-18 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.07 Å) | | Cite: | Conformational Activation Promotes CRISPR-Cas12a Catalysis and Resetting of the Endonuclease Activity.
Cell, 175, 2018
|
|
6GTF
 
 | | Transient state structure of CRISPR-Cpf1 (Cas12a) I5 conformation | | Descriptor: | CRISPR-associated endonuclease Cas12a, DNA (5'-D(P*AP*TP*GP*AP*CP*TP*TP*CP*TP*CP*TP*AP*AP*CP*AP*AP*GP*CP*TP*CP*G)-3'), DNA (5'-D(P*CP*GP*AP*GP*CP*TP*CP*GP*TP*TP*AP*GP*AP*GP*AP*A)-3'), ... | | Authors: | Montoya, G, Mesa, P, Stella, S. | | Deposit date: | 2018-06-18 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Conformational Activation Promotes CRISPR-Cas12a Catalysis and Resetting of the Endonuclease Activity.
Cell, 175, 2018
|
|
6GTD
 
 | | Transient state structure of CRISPR-Cpf1 (Cas12a) I2 conformation | | Descriptor: | CRISPR-associated endonuclease Cas12a, DNA (5'-D(P*CP*GP*AP*GP*CP*TP*CP*GP*TP*TP*AP*GP*AP*GP*AP*AP*GP*T)-3'), DNA (5'-D(P*TP*GP*AP*CP*TP*TP*CP*TP*CP*TP*AP*AP*CP*AP*AP*GP*CP*TP*CP*G)-3'), ... | | Authors: | Montoya, G, Mesa, P. | | Deposit date: | 2018-06-18 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.24 Å) | | Cite: | Conformational Activation Promotes CRISPR-Cas12a Catalysis and Resetting of the Endonuclease Activity.
Cell, 175, 2018
|
|
