8QJK
 
 | | Structure of the cytoplasmic domain of csx23 from Vibrio cholera in complex with cyclic tetra-adenylate (cA4) | | Descriptor: | ACETYL GROUP, Cyclic tetraadenosine monophosphate (cA4), SODIUM ION, ... | | Authors: | McMahon, S.A, McQuarrie, S, Gloster, T.M, Gruschow, S, White, M.F. | | Deposit date: | 2023-09-13 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.761 Å) | | Cite: | A cyclic-nucleotide binding membrane protein provides CRISPR-mediated antiphage defence in Vibrio cholera
To Be Published
|
|
6SCE
 
 | | Structure of a Type III CRISPR defence DNA nuclease activated by cyclic oligoadenylate | | Descriptor: | Uncharacterized protein, cyclic oligoadenylate | | Authors: | McMahon, S.A, Zhu, W, Graham, S, White, M.F, Gloster, T.M. | | Deposit date: | 2019-07-24 | | Release date: | 2020-02-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure and mechanism of a Type III CRISPR defence DNA nuclease activated by cyclic oligoadenylate.
Nat Commun, 11, 2020
|
|
6SCF
 
 | | A viral anti-CRISPR subverts type III CRISPR immunity by rapid degradation of cyclic oligoadenylate | | Descriptor: | Uncharacterized protein, cyclic oligoadenylate | | Authors: | McMahon, S.A, Athukoralage, J.S, Graham, S, White, M.F, Gloster, T.M. | | Deposit date: | 2019-07-24 | | Release date: | 2019-10-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | An anti-CRISPR viral ring nuclease subverts type III CRISPR immunity.
Nature, 577, 2020
|
|
3FFE
 
 | | Structure of Achromobactin Synthetase Protein D, (AcsD) | | Descriptor: | AcsD | | Authors: | McMahon, S.A, Liu, H, Carter, L, Oke, M, Johnson, K.A, Schmelz, S, Challis, G.L, White, M.F, Naismith, J.H, Scottish Structural Proteomics Facility (SSPF) | | Deposit date: | 2008-12-03 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | AcsD catalyzes enantioselective citrate desymmetrization in siderophore biosynthesis
Nat.Chem.Biol., 5, 2009
|
|
1YU1
 
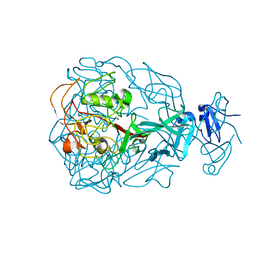 | | Major Tropism Determinant P3c Variant | | Descriptor: | MAGNESIUM ION, METHYL MERCURY ION, Major Tropism Determinant (Mtd-P3c) | | Authors: | McMahon, S.A, Miller, J.L, Lawton, J.A, Ghosh, P. | | Deposit date: | 2005-02-11 | | Release date: | 2005-09-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | The C-type lectin fold as an evolutionary solution for massive sequence variation
Nat.Struct.Mol.Biol., 12, 2005
|
|
1YU0
 
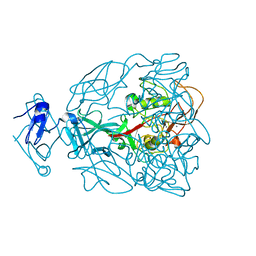 | | Major Tropism Determinant P1 Variant | | Descriptor: | CALCIUM ION, Major Tropism Determinant (Mtd-P1) | | Authors: | McMahon, S.A, Miller, J.L, Lawton, J.A, Ghosh, P. | | Deposit date: | 2005-02-11 | | Release date: | 2005-09-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | The C-type lectin fold as an evolutionary solution for massive sequence variation
Nat.Struct.Mol.Biol., 12, 2005
|
|
1YU2
 
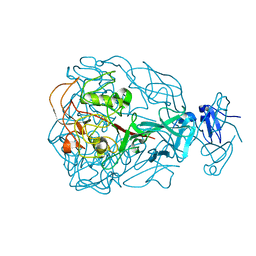 | | Major Tropism Determinant M1 Variant | | Descriptor: | MAGNESIUM ION, Major Tropism Determinant (Mtd-M1) | | Authors: | McMahon, S.A, Miller, J.L, Lawton, J.A, Ghosh, P. | | Deposit date: | 2005-02-11 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The C-type lectin fold as an evolutionary solution for massive sequence variation
Nat.Struct.Mol.Biol., 12, 2005
|
|
1YU3
 
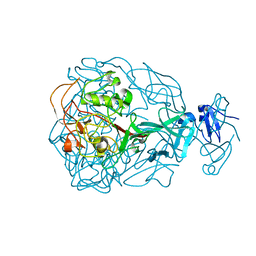 | | Major Tropism Determinant I1 Variant | | Descriptor: | MAGNESIUM ION, Major Tropism Determinant (Mtd-I1) | | Authors: | McMahon, S.A, Miller, J.L, Lawton, J.A, Ghosh, P. | | Deposit date: | 2005-02-11 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | The C-type lectin fold as an evolutionary solution for massive sequence variation
Nat.Struct.Mol.Biol., 12, 2005
|
|
1YU4
 
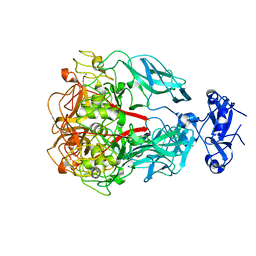 | | Major Tropism Determinant U1 Variant | | Descriptor: | MAGNESIUM ION, Major Tropism Determinant (Mtd-U1) | | Authors: | McMahon, S.A, Miller, J.L, Lawton, J.A, Ghosh, P. | | Deposit date: | 2005-02-11 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The C-type lectin fold as an evolutionary solution for massive sequence variation
Nat.Struct.Mol.Biol., 12, 2005
|
|
2WR8
 
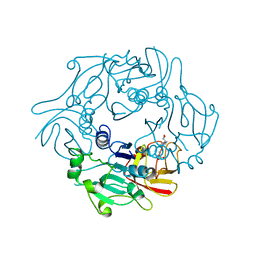 | | Structure of Pyrococcus horikoshii SAM hydroxide adenosyltransferase in complex with SAH | | Descriptor: | PUTATIVE UNCHARACTERIZED PROTEIN PH0463, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | McMahon, S.A, Deng, H, O'Hagan, D, Johnson, K.A, Naismith, J.H. | | Deposit date: | 2009-08-31 | | Release date: | 2009-09-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Mechanistic Insights Into Water Activation in Sam Hydroxide Adenosyltransferase (Duf-62).
Chembiochem, 10, 2009
|
|
4BQQ
 
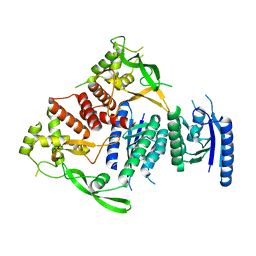 | |
4AVF
 
 | | Crystal structure of Pseudomonas aeruginosa inosine 5'-monophosphate dehydrogenase | | Descriptor: | INOSINE-5'-MONOPHOSPHATE DEHYDROGENASE | | Authors: | McMahon, S.A, Moynie, L, Liu, H, Duthie, F, Naismith, J.H. | | Deposit date: | 2012-05-25 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | The Aeropath Project Targeting Pseudomonas Aeruginosa: Crystallographic Studies for Assessment of Potential Targets in Early-Stage Drug Discovery
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4CQJ
 
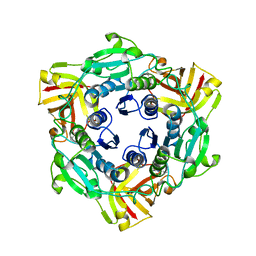 | |
4AVR
 
 | | Crystal structure of the hypothetical protein Pa4485 from Pseudomonas aeruginosa | | Descriptor: | PA4485 | | Authors: | McMahon, S.A, Moynie, L, Liu, H, Duthie, F, Alphey, M.S, Naismith, J.H. | | Deposit date: | 2012-05-29 | | Release date: | 2013-01-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | The Aeropath Project Targeting Pseudomonas Aeruginosa: Crystallographic Studies for Assessment of Potential Targets in Early-Stage Drug Discovery.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4CCV
 
 | | Crystal structure of histidine-rich glycoprotein N2 domain reveals redox activity at an interdomain disulfide bridge: Implications for the regulation of angiogenesis | | Descriptor: | GLUTATHIONE, GLYCEROL, HISTIDINE-RICH GLYCOPROTEIN, ... | | Authors: | McMahon, S.A, Kassaar, O, Stewart, A.J, Naismith, J.H. | | Deposit date: | 2013-10-29 | | Release date: | 2014-02-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal Structure of Histidine-Rich Glycoprotein N2 Domain Reveals Redox Activity at an Interdomain Disulfide Bridge: Implications for Angiogenic Regulation.
Blood, 123, 2014
|
|
2W82
 
 | | The structure of ArdA | | Descriptor: | ORF18 | | Authors: | McMahon, S.A, Roberts, G.A, Carter, L.G, Cooper, L.P, Liu, H, White, J.H, Johnson, K.A, Sanghvi, B, Oke, M, Walkinshaw, M.D, Blakely, G, Naismith, J.H, Dryden, D.T.F. | | Deposit date: | 2009-01-08 | | Release date: | 2009-01-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Extensive DNA Mimicry by the Arda Anti-Restriction Protein and its Role in the Spread of Antibiotic Resistance.
Nucleic Acids Res., 37, 2009
|
|
6ENK
 
 | | The X-ray crystal structure of DesE bound to desferrioxamine B | | Descriptor: | DesE, SODIUM ION, desferrioxamine B | | Authors: | Naismith, J.H, McMahon, S.A, Challis, G.L, Kadi, N, Oke, M, Liu, H, Carter, L.G, Johnson, K.A. | | Deposit date: | 2017-10-05 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Desferrioxamine biosynthesis: diverse hydroxamate assembly by substrate-tolerant acyl transferase DesC.
Philos. Trans. R. Soc. Lond., B, Biol. Sci., 373, 2018
|
|
6I6X
 
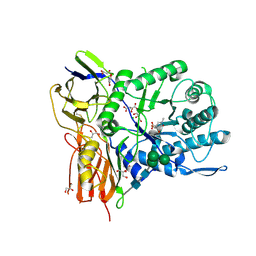 | | New Irreversible a-l-Iduronidase Inhibitors and Activity-Based Probes | | Descriptor: | (1~{R},2~{R},3~{R},4~{S},5~{S},6~{R})-7-methyl-3,4,5-tris(oxidanyl)-7-azabicyclo[4.1.0]heptane-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gloster, T.M, McMahon, S.A, Oehler, V. | | Deposit date: | 2018-11-15 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | New Irreversible alpha-l-Iduronidase Inhibitors and Activity-Based Probes.
Chemistry, 24, 2018
|
|
6I6R
 
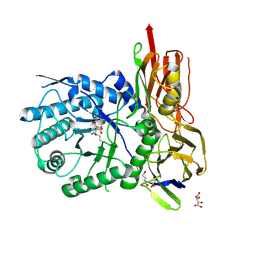 | | New Irreversible a-l-Iduronidase Inhibitors and Activity-Based Probes | | Descriptor: | (1~{R},2~{R},3~{R},4~{S},6~{S})-6-azanyl-2,3,4-tris(oxidanyl)cyclohexane-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-L-iduronidase, ... | | Authors: | Gloster, T.M, McMahon, S.A, Oehler, V. | | Deposit date: | 2018-11-15 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | New Irreversible alpha-l-Iduronidase Inhibitors and Activity-Based Probes.
Chemistry, 24, 2018
|
|
8PCW
 
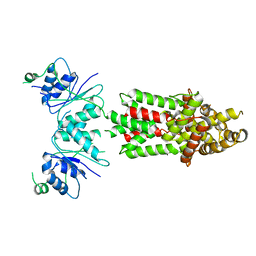 | | Structure of Csm6' from Streptococcus thermophilus | | Descriptor: | CRISPR system endoribonuclease Csm6' | | Authors: | McQuarrie, S.J, Athukoralage, J.S, McMahon, S.A, Graham, S, Ackerman, K, Bode, B.E, White, M.F, Gloster, T.M. | | Deposit date: | 2023-06-11 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.54 Å) | | Cite: | Activation of Csm6 ribonuclease by cyclic nucleotide binding: in an emergency, twist to open.
Nucleic Acids Res., 51, 2023
|
|
8PE3
 
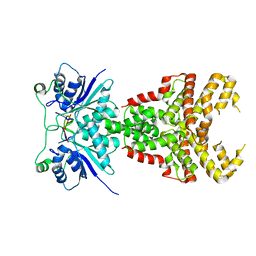 | | Structure of Csm6' from Streptococcus thermophilus in complex with cyclic hexa-adenylate (cA6) | | Descriptor: | CRISPR system endoribonuclease Csm6', Cyclic hexaadenosine monophosphate (cA6), RNA | | Authors: | McQuarrie, S.J, Athukoralage, J.S, McMahon, S.A, Graham, S, Ackerman, K, Bode, B.E, White, M.F, Gloster, T.M. | | Deposit date: | 2023-06-13 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Activation of Csm6 ribonuclease by cyclic nucleotide binding: in an emergency, twist to open.
Nucleic Acids Res., 51, 2023
|
|
6ZZP
 
 | | Crystal structure of (R)-3-hydroxybutyrate dehydrogenase from Psychrobacter arcticus complexed with NAD+ and 3-oxovalerate | | Descriptor: | 3-oxidanylidenepentanoic acid, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative beta-hydroxybutyrate dehydrogenase | | Authors: | Machado, T.F.G, da Silva, R.G, Gloster, T.M, McMahon, S.A, Oehler, V. | | Deposit date: | 2020-08-04 | | Release date: | 2020-10-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Dissecting the Mechanism of ( R )-3-Hydroxybutyrate Dehydrogenase by Kinetic Isotope Effects, Protein Crystallography, and Computational Chemistry.
Acs Catalysis, 10, 2020
|
|
6ZZQ
 
 | | Crystal structure of (R)-3-hydroxybutyrate dehydrogenase from Acinetobacter baumannii complexed with NAD+ and acetoacetate | | Descriptor: | 3-hydroxybutyrate dehydrogenase, ACETOACETIC ACID, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Machado, T.F.G, da Silva, R.G, Gloster, T.M, McMahon, S.A, Oehler, V. | | Deposit date: | 2020-08-05 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Dissecting the Mechanism of ( R )-3-Hydroxybutyrate Dehydrogenase by Kinetic Isotope Effects, Protein Crystallography, and Computational Chemistry.
Acs Catalysis, 10, 2020
|
|
5FIU
 
 | | Binding and structural studies of a 5,5-difluoromethyl adenosine nucleoside with the fluorinase enzyme | | Descriptor: | 5'-FLUORO-5'-DEOXY-ADENOSINE SYNTHASE, 5,5-DIFLUOROMETHYL ADENOSINE, L(+)-TARTARIC ACID | | Authors: | Thompson, S, McMahon, S.A, Naismith, J.H, O'Hagan, D. | | Deposit date: | 2015-10-02 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Exploration of a Potential Difluoromethyl-Nucleoside Substrate with the Fluorinase Enzyme.
Bioorg.Chem., 64, 2015
|
|
7BDV
 
 | | Structure of Can2 from Sulfobacillus thermosulfidooxidans in complex with cyclic tetra-adenylate (cA4) | | Descriptor: | Can2, Cyclic tetraadenosine monophosphate (cA4) | | Authors: | McQuarrie, S, McMahon, S.A, Gloster, T.M, White, M.F, Graham, S, Zhu, W, Gruschow, S. | | Deposit date: | 2020-12-22 | | Release date: | 2021-03-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The CRISPR ancillary effector Can2 is a dual-specificity nuclease potentiating type III CRISPR defence.
Nucleic Acids Res., 49, 2021
|
|
