2K9Y
 
 | |
3GFC
 
 | | Crystal Structure of Histone-binding protein RBBP4 | | Descriptor: | Histone-binding protein RBBP4 | | Authors: | Amaya, M.F, Dong, A, Li, Z, He, H, Ni, S, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-02-26 | | Release date: | 2009-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and function of WD40 domain proteins.
Protein Cell, 2, 2011
|
|
2OJT
 
 | | Structure and mechanism of kainate receptor modulation by anions | | Descriptor: | (S)-1-(2-AMINO-2-CARBOXYETHYL)-3(2-CARBOXYTHIOPHENE-3-YL-METHYL)-5-METHYLPYRIMIDINE-2,4-DIONE, BROMIDE ION, Glutamate receptor, ... | | Authors: | Mayer, M.L. | | Deposit date: | 2007-01-14 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and mechanism of kainate receptor modulation by anions.
Neuron, 53, 2007
|
|
3I2N
 
 | | Crystal Structure of WD40 repeats protein WDR92 | | Descriptor: | WD repeat-containing protein 92 | | Authors: | Amaya, M.F, Li, Z, He, H, Seitova, A, Ni, S, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-06-29 | | Release date: | 2009-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and function of WD40 domain proteins.
Protein Cell, 2, 2011
|
|
1SD3
 
 | |
1S50
 
 | |
1S9T
 
 | |
1S7Y
 
 | |
1TXF
 
 | |
1TT1
 
 | |
2AH2
 
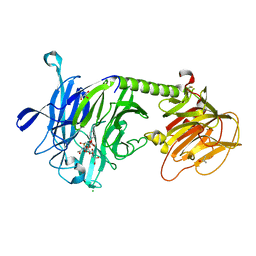 | | Trypanosoma cruzi trans-sialidase in complex with 2,3-difluorosialic acid (covalent intermediate) | | Descriptor: | 5-acetamido-3,5-dideoxy-3-fluoro-D-erythro-alpha-L-manno-non-2-ulopyranosonic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Amaya, M.F, Watts, A.G, Damager, I, Wehenkel, A, Nguyen, T, Buschiazzo, A, Paris, G, Frasch, A.C, Withers, S.G, Alzari, P.M. | | Deposit date: | 2005-07-27 | | Release date: | 2005-08-23 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Insights into the Catalytic Mechanism of Trypanosoma cruzi trans-Sialidase
Structure, 12, 2004
|
|
2A75
 
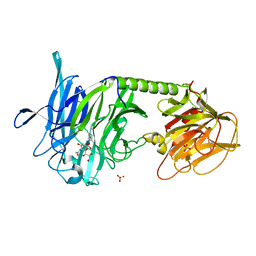 | | Trypanosoma rangeli Sialidase In Complex With 2,3- Difluorosialic Acid (Covalent Intermediate) | | Descriptor: | 5-acetamido-3,5-dideoxy-3-fluoro-D-erythro-alpha-L-manno-non-2-ulopyranosonic acid, SULFATE ION, sialidase | | Authors: | Amaya, M.F, Alzari, P.M, Buschiazzo, A. | | Deposit date: | 2005-07-04 | | Release date: | 2005-11-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and Kinetic Analysis of Two Covalent Sialosyl-Enzyme Intermediates on Trypanosoma rangeli Sialidase.
J.Biol.Chem., 281, 2006
|
|
2AGS
 
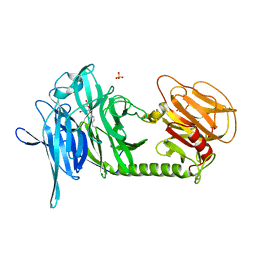 | | Trypanosoma rangeli Sialidase in Complex with 2-Keto-3-deoxy-D-glycero-D-galacto-2,3-difluoro-nononic acid (2,3-difluoro-KDN) | | Descriptor: | 3-deoxy-3-fluoro-D-erythro-alpha-L-manno-non-2-ulopyranosonic acid, SULFATE ION, sialidase | | Authors: | Amaya, M.F, Alzari, P.M, Buschiazzo, A. | | Deposit date: | 2005-07-27 | | Release date: | 2005-11-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Kinetic Analysis of Two Covalent Sialosyl-Enzyme Intermediates on Trypanosoma rangeli Sialidase.
J.Biol.Chem., 281, 2006
|
|
4UQQ
 
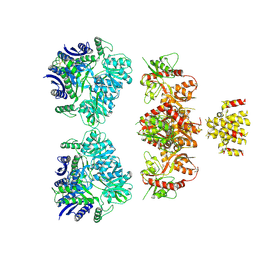 | | Electron density map of GluK2 desensitized state in complex with 2S,4R-4-methylglutamate | | Descriptor: | GLUTAMATE RECEPTOR IONOTROPIC, KAINATE 2, GLUTAMIC ACID | | Authors: | Meyerson, J.R, Kumar, J, Chittori, S, Rao, P, Pierson, J, Bartesaghi, A, Mayer, M.L, Subramaniam, S. | | Deposit date: | 2014-06-24 | | Release date: | 2014-08-13 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Structural Mechanism of Glutamate Receptor Activation and Desensitization
Nature, 514, 2014
|
|
4UQJ
 
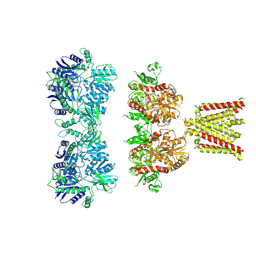 | | Cryo-EM density map of GluA2em in complex with ZK200775 | | Descriptor: | GLUTAMATE RECEPTOR 2, {[7-morpholin-4-yl-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl]methyl}phosphonic acid | | Authors: | Meyerson, J.R, Kumar, J, Chittori, S, Rao, P, Pierson, J, Bartesaghi, A, Mayer, M.L, Subramaniam, S. | | Deposit date: | 2014-06-24 | | Release date: | 2014-08-13 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (10.4 Å) | | Cite: | Structural Mechanism of Glutamate Receptor Activation and Desensitization
Nature, 514, 2014
|
|
6NJF
 
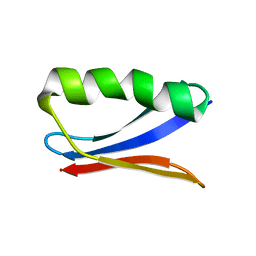 | | Solution NMR Structure of DANCER3-F34A, a rigid and natively folded single mutant of the dynamic protein DANCER-3 | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Damry, A.M, Mayer, M.M, Goto, N.K, Chica, R.A. | | Deposit date: | 2019-01-03 | | Release date: | 2019-08-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Origin of conformational dynamics in a globular protein.
Commun Biol, 2, 2019
|
|
6OAE
 
 | |
8D40
 
 | |
4UQK
 
 | | Electron density map of GluA2em in complex with quisqualate and LY451646 | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, GLUTAMATE RECEPTOR 2 | | Authors: | Meyerson, J.R, Kumar, J, Chittori, S, Rao, P, Pierson, J, Bartesaghi, A, Mayer, M.L, Subramaniam, S. | | Deposit date: | 2014-06-24 | | Release date: | 2014-08-13 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (16.4 Å) | | Cite: | Structural Mechanism of Glutamate Receptor Activation and Desensitization
Nature, 514, 2014
|
|
5CMK
 
 | | Crystal structure of the GluK2EM LBD dimer assembly complex with glutamate and LY466195 | | Descriptor: | (3S,4aR,6S,8aR)-6-{[(2S)-2-carboxy-4,4-difluoropyrrolidin-1-yl]methyl}decahydroisoquinoline-3-carboxylic acid, CHLORIDE ION, GLUTAMIC ACID, ... | | Authors: | Chittori, S, Mayer, M.L. | | Deposit date: | 2015-07-16 | | Release date: | 2016-07-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural basis of kainate subtype glutamate receptor desensitization.
Nature, 537, 2016
|
|
5CMM
 
 | |
9CUO
 
 | | Crystal structure of CRBN with compound 3 | | Descriptor: | (3S)-3-(3-methyl-2-oxo-2,3-dihydro-1H-1,3-benzimidazol-1-yl)piperidine-2,6-dione, 1,2-ETHANEDIOL, Protein cereblon, ... | | Authors: | Zheng, X, Ji, N, Campbell, V, Slavin, A, Zhu, X, Chen, D, Rong, H, Enerson, B, Mayo, M, Sharma, K, Browne, C.M, Klaus, C.R, Li, H, Massa, G, McDonald, A.A, Shi, Y, Sintchak, M, Skouras, S, Walther, D.M, Yuan, K, Zhang, Y, Kelleher, J, Guang, L, Luo, X, Mainolfi, N, Weiss, M.M. | | Deposit date: | 2024-07-26 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of KT-474─a Potent, Selective, and Orally Bioavailable IRAK4 Degrader for the Treatment of Autoimmune Diseases.
J.Med.Chem., 2024
|
|
5CO1
 
 | |
6E8F
 
 | |
1V9U
 
 | | Human Rhinovirus 2 bound to a fragment of its cellular receptor protein | | Descriptor: | CALCIUM ION, Coat protein VP1, Coat protein VP2, ... | | Authors: | Verdaguer, N, Fita, I, Reithmayer, M, Moser, R, Blaas, D. | | Deposit date: | 2004-02-03 | | Release date: | 2004-05-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | X-ray structure of a minor group human rhinovirus bound to a fragment of its cellular receptor protein
NAT.STRUCT.MOL.BIOL., 11, 2004
|
|
