1MS1
 
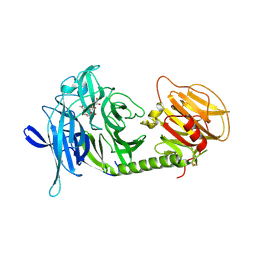 | | Monoclinic form of Trypanosoma cruzi trans-sialidase, in complex with 3-deoxy-2,3-dehydro-N-acetylneuraminic acid (DANA) | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, GLYCEROL, trans-sialidase | | Authors: | Buschiazzo, A, Amaya, M.F, Cremona, M.L, Frasch, A.C, Alzari, P.M. | | Deposit date: | 2002-09-19 | | Release date: | 2003-03-25 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure and mode of action of trans-sialidase, a key enzyme in Trypanosoma cruzi pathogenesis
Mol.Cell, 10, 2002
|
|
1MR5
 
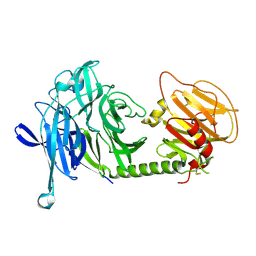 | | Orthorhombic form of Trypanosoma cruzi trans-sialidase | | Descriptor: | trans-sialidase | | Authors: | Buschiazzo, A, Amaya, M.F, Cremona, M.L, Frasch, A.C, Alzari, P.M. | | Deposit date: | 2002-09-18 | | Release date: | 2003-03-18 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The crystal structure and mode of action of trans-sialidase, a key enzyme in Trypanosoma cruzi pathogenesis
Mol.Cell, 10, 2002
|
|
1MS8
 
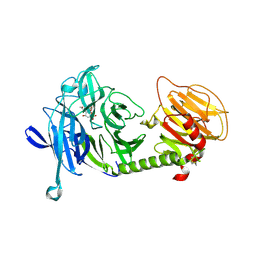 | | Triclinic form of Trypanosoma cruzi trans-sialidase, in complex with 3-deoxy-2,3-dehydro-N-acetylneuraminic acid (DANA) | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, trans-sialidase | | Authors: | Buschiazzo, A, Amaya, M.F, Cremona, M.L, Frasch, A.C, Alzari, P.M. | | Deposit date: | 2002-09-19 | | Release date: | 2003-03-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure and mode of action of trans-sialidase, a key enzyme in Trypanosoma cruzi pathogenesis
Mol.Cell, 10, 2002
|
|
1SS3
 
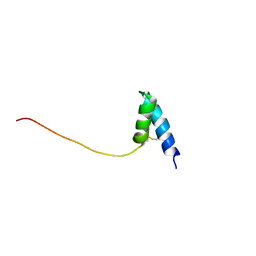 | | Solution structure of Ole e 6, an allergen from olive tree pollen | | Descriptor: | Pollen allergen Ole e 6 | | Authors: | Trevino, M.A, Garcia-Mayoral, M.F, Barral, P, Villalba, M, Santoro, J, Rico, M, Rodriguez, R, Bruix, M. | | Deposit date: | 2004-03-23 | | Release date: | 2004-08-03 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of Ole e 6, a Major Allergen from Olive Tree Pollen.
J.Biol.Chem., 279, 2004
|
|
1MS9
 
 | | Triclinic form of Trypanosoma cruzi trans-sialidase, in complex with lactose | | Descriptor: | beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, trans-sialidase | | Authors: | Buschiazzo, A, Amaya, M.F, Cremona, M.L, Frasch, A.C, Alzari, P.M. | | Deposit date: | 2002-09-19 | | Release date: | 2003-03-25 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The crystal structure and mode of action of trans-sialidase, a key enzyme of Trypanosoma cruzi pathogenesis
Mol.Cell, 10, 2002
|
|
1R4Y
 
 | | SOLUTION STRUCTURE OF THE DELETION MUTANT DELTA(7-22) OF THE CYTOTOXIC RIBONUCLEASE ALPHA-SARCIN | | Descriptor: | Ribonuclease alpha-sarcin | | Authors: | Garcia-Mayoral, M.F, Garcia-Ortega, L, Lillo, M.P, Santoro, J, Martinez Del Pozo, A, Gavilanes, J.G, Rico, M, Bruix, M. | | Deposit date: | 2003-10-09 | | Release date: | 2004-04-06 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the noncytotoxic {alpha}-sarcin mutant {Delta}(7-22): The importance of the native conformation of peripheral loops for activity.
Protein Sci., 13, 2004
|
|
2M7L
 
 | |
1MQJ
 
 | | Crystal structure of the GluR2 ligand binding core (S1S2J) in complex with willardiine at 1.65 angstroms resolution | | Descriptor: | 2-AMINO-3-(2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, ZINC ION, glutamate receptor 2 | | Authors: | Jin, R, Banke, T.G, Mayer, M.L, Traynelis, S.F, Gouaux, E. | | Deposit date: | 2002-09-16 | | Release date: | 2003-08-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for partial agonist action at ionotropic glutamate receptors
Nat.Neurosci., 6, 2003
|
|
1R7S
 
 | | PUTIDAREDOXIN (Fe2S2 ferredoxin), C73G mutant | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Putidaredoxin | | Authors: | Smith, N, Mayhew, M, Kelly, H, Robinson, H, Heroux, A, Holden, M.J, Gallagher, D.T. | | Deposit date: | 2003-10-22 | | Release date: | 2004-04-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure of C73G putidaredoxin from Pseudomonas putida.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1MQG
 
 | | Crystal Structure of the GluR2 Ligand Binding Core (S1S2J) in Complex with Iodo-Willardiine at 2.15 Angstroms Resolution | | Descriptor: | 2-AMINO-3-(5-IODO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Jin, R, Banke, T.G, Mayer, M.L, Traynelis, S.F, Gouaux, E. | | Deposit date: | 2002-09-16 | | Release date: | 2003-08-05 | | Last modified: | 2017-05-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for partial agonist action at ionotropic glutamate receptors
Nat.Neurosci., 6, 2003
|
|
1LB8
 
 | | Crystal structure of the Non-desensitizing GluR2 ligand binding core mutant (S1S2J-L483Y) in complex with AMPA at 2.3 resolution | | Descriptor: | (S)-ALPHA-AMINO-3-HYDROXY-5-METHYL-4-ISOXAZOLEPROPIONIC ACID, Glutamate receptor 2 | | Authors: | Sun, Y, Olson, R, Horning, M, Armstrong, N, Mayer, M, Gouaux, E. | | Deposit date: | 2002-04-02 | | Release date: | 2002-06-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanism of glutamate receptor desensitization.
Nature, 417, 2002
|
|
1MQI
 
 | | Crystal Structure of the GluR2 Ligand Binding Core (S1S2J) in Complex with Fluoro-Willardiine at 1.35 Angstroms Resolution | | Descriptor: | 2-AMINO-3-(5-FLUORO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, glutamate receptor 2 | | Authors: | Jin, R, Banke, T.G, Mayer, M.L, Traynelis, S.F, Gouaux, E. | | Deposit date: | 2002-09-16 | | Release date: | 2003-08-05 | | Last modified: | 2017-08-02 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural basis for partial agonist action at ionotropic glutamate receptors
Nat.Neurosci., 6, 2003
|
|
1LBB
 
 | | Crystal structure of the GluR2 ligand binding domain mutant (S1S2J-N754D) in complex with kainate at 2.1 A resolution | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamine receptor 2 | | Authors: | Sun, Y, Olson, R, Horning, M, Armstrong, N, Mayer, M, Gouaux, E. | | Deposit date: | 2002-04-02 | | Release date: | 2002-06-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of glutamate receptor desensitization.
Nature, 417, 2002
|
|
1LB9
 
 | | Crystal structure of the Non-desensitizing GluR2 ligand binding core mutant (S1S2J-L483Y) in complex with antagonist DNQX at 2.3 A resolution | | Descriptor: | 6,7-DINITROQUINOXALINE-2,3-DIONE, Glutamate receptor 2, SULFATE ION | | Authors: | Sun, Y, Olson, R, Horning, M, Armstrong, N, Mayer, M, Gouaux, E. | | Deposit date: | 2002-04-02 | | Release date: | 2002-06-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanism of glutamate receptor desensitization.
Nature, 417, 2002
|
|
1P1N
 
 | |
1P1O
 
 | | Crystal structure of the GluR2 ligand-binding core (S1S2J) mutant L650T in complex with quisqualate | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, Glutamate receptor 2, SULFATE ION | | Authors: | Armstrong, N, Mayer, M.L, Gouaux, E. | | Deposit date: | 2003-04-13 | | Release date: | 2003-06-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Tuning activation of the AMPA-sensitive GluR2 ion channel by genetic
adjustment of agonist-induced conformational changes.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1MQH
 
 | | Crystal Structure of the GluR2 Ligand Binding Core (S1S2J) in Complex with Bromo-Willardiine at 1.8 Angstroms Resolution | | Descriptor: | 2-AMINO-3-(5-BROMO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, glutamate receptor 2 | | Authors: | Jin, R, Banke, T.G, Mayer, M.L, Traynelis, S.F, Gouaux, E. | | Deposit date: | 2002-09-16 | | Release date: | 2003-08-05 | | Last modified: | 2017-08-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for partial agonist action at ionotropic glutamate receptors
Nat.Neurosci., 6, 2003
|
|
1RU5
 
 | |
1P1W
 
 | |
1P1Q
 
 | |
1P1U
 
 | |
1RUU
 
 | |
1SEN
 
 | | Endoplasmic reticulum protein Rp19 O95881 | | Descriptor: | CHLORIDE ION, PLATINUM (II) ION, Thioredoxin-like protein p19 | | Authors: | Liu, Z.-J, Chen, L, Tempel, W, Shah, A, Lee, D, Dailey, T.A, Mayer, M.R, Rose, J.P, Richardson, D.C, Richardson, J.S, Dailey, H.A, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-02-17 | | Release date: | 2004-07-13 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.199 Å) | | Cite: | Endoplasmic reticulum protein Rp19
To be Published
|
|
