5GLK
 
 | | Crystal structure of CoXyl43, GH43 beta-xylosidase/alpha-arabinofuranosidase from a compost microbial metagenome, calcium-free form. | | Descriptor: | ACETATE ION, GLYCEROL, Glycoside hydrolase family 43, ... | | Authors: | Matsuzawa, T, Kishine, N, Fujimoto, Z, Yaoi, K. | | Deposit date: | 2016-07-12 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of metagenomic beta-xylosidase/ alpha-l-arabinofuranosidase activated by calcium.
J. Biochem., 162, 2017
|
|
5GLO
 
 | | Crystal structure of CoXyl43, GH43 beta-xylosidase/alpha-arabinofuranosidase from a compostmicrobial metagenome in complex with l-arabinose, calcium-free form | | Descriptor: | ACETATE ION, Glycoside hydrolase family 43, SODIUM ION, ... | | Authors: | Matsuzawa, T, Kishine, N, Fujimoto, Z, Yaoi, K. | | Deposit date: | 2016-07-12 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of metagenomic beta-xylosidase/ alpha-l-arabinofuranosidase activated by calcium.
J. Biochem., 162, 2017
|
|
5GLQ
 
 | | Crystal structure of CoXyl43, GH43 beta-xylosidase/alpha-arabinofuranosidase from a compostmicrobial metagenome in complex with l-arabinose and xylotriose, calcium-free form | | Descriptor: | Glycoside hydrolase family 43, SODIUM ION, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, ... | | Authors: | Matsuzawa, T, Kishine, N, Fujimoto, Z, Yaoi, K. | | Deposit date: | 2016-07-12 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of metagenomic beta-xylosidase/ alpha-l-arabinofuranosidase activated by calcium.
J. Biochem., 162, 2017
|
|
5GLR
 
 | | Crystal structure of CoXyl43, GH43 beta-xylosidase/alpha-arabinofuranosidase from a compostmicrobial metagenome in complex with l-arabinose and xylotriose, calcium-bound form | | Descriptor: | CALCIUM ION, Glycoside hydrolase family 43, SODIUM ION, ... | | Authors: | Matsuzawa, T, Kishine, N, Fujimoto, Z, Yaoi, K. | | Deposit date: | 2016-07-12 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of metagenomic beta-xylosidase/ alpha-l-arabinofuranosidase activated by calcium.
J. Biochem., 162, 2017
|
|
5GLL
 
 | | Crystal structure of CoXyl43, GH43 beta-xylosidase/alpha-arabinofuranosidase from a compostmicrobial metagenome, calcium-bound form | | Descriptor: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Matsuzawa, T, Kishine, N, Fujimoto, Z, Yaoi, K. | | Deposit date: | 2016-07-12 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of metagenomic beta-xylosidase/ alpha-l-arabinofuranosidase activated by calcium.
J. Biochem., 162, 2017
|
|
5GLN
 
 | | Crystal structure of CoXyl43, GH43 beta-xylosidase/alpha-arabinofuranosidase from a compostmicrobial metagenome in complex with xylotriose, calcium-bound form | | Descriptor: | ACETATE ION, CALCIUM ION, Glycoside hydrolase family 43, ... | | Authors: | Matsuzawa, T, Kishine, N, Fujimoto, Z, Yaoi, K. | | Deposit date: | 2016-07-12 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of metagenomic beta-xylosidase/ alpha-l-arabinofuranosidase activated by calcium.
J. Biochem., 162, 2017
|
|
5GLP
 
 | | Crystal structure of CoXyl43, GH43 beta-xylosidase/alpha-arabinofuranosidase from a compostmicrobial metagenome in complex with l-arabinose, calcium-bound form | | Descriptor: | ACETATE ION, CALCIUM ION, Glycoside hydrolase family 43, ... | | Authors: | Matsuzawa, T, Kishine, N, Fujimoto, Z, Yaoi, K. | | Deposit date: | 2016-07-12 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of metagenomic beta-xylosidase/ alpha-l-arabinofuranosidase activated by calcium.
J. Biochem., 162, 2017
|
|
5GLM
 
 | | Crystal structure of CoXyl43, GH43 beta-xylosidase/alpha-arabinofuranosidase from a compost microbial metagenome in complex with xylotriose, calcium-free form. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, Glycoside hydrolase family 43, ... | | Authors: | Matsuzawa, T, Kishine, N, Fujimoto, Z, Yaoi, K. | | Deposit date: | 2016-07-12 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of metagenomic beta-xylosidase/ alpha-l-arabinofuranosidase activated by calcium.
J. Biochem., 162, 2017
|
|
3WVR
 
 | | Structure of ATP grasp protein with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, PGM1, ... | | Authors: | Matsui, T, Noike, M, Ooya, K, Sasaki, I, Hamano, Y, Maruyama, C, Ishikawa, J, Satoh, Y, Ito, H, Dairi, T, Morita, H. | | Deposit date: | 2014-06-04 | | Release date: | 2014-11-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.175 Å) | | Cite: | A peptide ligase and the ribosome cooperate to synthesize the peptide pheganomycin.
Nat.Chem.Biol., 11, 2015
|
|
3WVQ
 
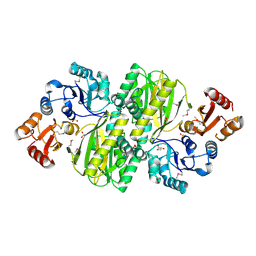 | | Structure of ATP grasp protein | | Descriptor: | GLYCEROL, PGM1, SULFATE ION | | Authors: | Matsui, T, Noike, M, Ooya, K, Sasaki, I, Hamano, Y, Maruyama, C, Ishikawa, J, Satoh, Y, Ito, H, Dairi, T, Morita, H. | | Deposit date: | 2014-06-04 | | Release date: | 2014-11-19 | | Last modified: | 2014-12-31 | | Method: | X-RAY DIFFRACTION (1.955 Å) | | Cite: | A peptide ligase and the ribosome cooperate to synthesize the peptide pheganomycin.
Nat.Chem.Biol., 11, 2015
|
|
5YOT
 
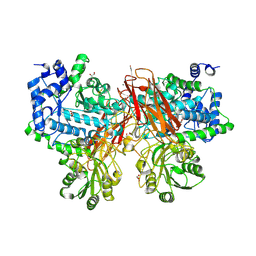 | | Isoprimeverose-producing enzyme from Aspergillus oryzae in complex with isoprimeverose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Matsuzawa, T, Watanabe, M, Nakamichi, Y, Yaoi, K. | | Deposit date: | 2017-10-31 | | Release date: | 2018-11-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structure and substrate recognition mechanism of Aspergillus oryzae isoprimeverose-producing enzyme.
J.Struct.Biol., 205, 2019
|
|
5ZN6
 
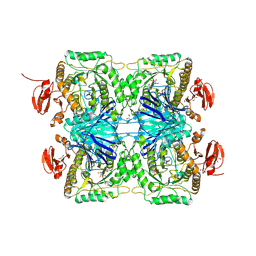 | |
5ZN7
 
 | |
2Z9S
 
 | | Crystal Structure Analysis of rat HBP23/Peroxiredoxin I, Cys52Ser mutant | | Descriptor: | Peroxiredoxin-1 | | Authors: | Matsumura, T, Okamoto, K, Nishino, T, Abe, Y. | | Deposit date: | 2007-09-25 | | Release date: | 2007-11-20 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Dimer-Oligomer Interconversion of Wild-type and Mutant Rat 2-Cys Peroxiredoxin: DISULFIDE FORMATION AT DIMER-DIMER INTERFACES IS NOT ESSENTIAL FOR DECAMERIZATION
J.Biol.Chem., 283, 2008
|
|
2DI2
 
 | | NMR structure of the HIV-2 nucleocapsid protein | | Descriptor: | Nucleocapsid protein p7, ZINC ION | | Authors: | Matsui, T, Kodera, Y, Endoh, H, Miyauchi, E, Komatsu, H, Sato, K, Tanaka, T, Kohno, T, Maeda, T. | | Deposit date: | 2006-03-27 | | Release date: | 2007-03-13 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | RNA Recognition Mechanism of the Minimal Active Domain of the Human Immunodeficiency Virus Type-2 Nucleocapsid Protein
J.Biochem.(Tokyo), 141, 2007
|
|
2E1X
 
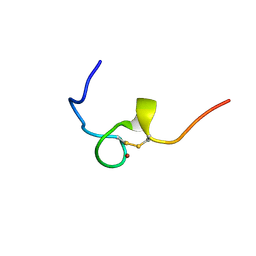 | | NMR structure of the HIV-2 nucleocapsid protein | | Descriptor: | Gag-Pol polyprotein (Pr160Gag-Pol), ZINC ION | | Authors: | Matsui, T, Kodera, Y, Miyauchi, E, Tanaka, H, Endoh, H, Komatsu, H, Tanaka, T, Kohno, T, Maeda, T. | | Deposit date: | 2006-11-03 | | Release date: | 2007-06-05 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structural role of the secondary active domain of HIV-2 NCp8 in multi-functionality
Biochem.Biophys.Res.Commun., 358, 2007
|
|
2EC7
 
 | | Solution Structure of Human Immunodificiency Virus Type-2 Nucleocapsid Protein | | Descriptor: | Gag polyprotein (Pr55Gag), ZINC ION | | Authors: | Matsui, T, Kodera, Y, Tanaka, T, Endoh, H, Tanaka, H, Miyauchi, E, Komatsu, H, Kohno, T, Maeda, T. | | Deposit date: | 2007-02-10 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The RNA recognition mechanism of human immunodeficiency virus (HIV) type 2 NCp8 is different from that of HIV-1 NCp7
Biochemistry, 48, 2009
|
|
6KO7
 
 | | Crystal structure of the Ethidium bound RamR determined with XtaLAB Synergy | | Descriptor: | ETHIDIUM, Putative regulatory protein, SULFATE ION | | Authors: | Matsumoto, T, Nakashima, R, Yamano, A, Nishino, K. | | Deposit date: | 2019-08-08 | | Release date: | 2019-10-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Development of a structure determination method using a multidrug-resistance regulator protein as a framework.
Biochem.Biophys.Res.Commun., 518, 2019
|
|
6KO9
 
 | | Crystal structure of the Gefitinib Intermediate 1 bound RamR determined with XtaLAB Synergy | | Descriptor: | 4-[(3-chloranyl-4-fluoranyl-phenyl)amino]-7-methoxy-quinazolin-6-ol, Putative regulatory protein, SULFATE ION | | Authors: | Matsumoto, T, Nakashima, R, Yamano, A, Nishino, K. | | Deposit date: | 2019-08-08 | | Release date: | 2019-10-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Development of a structure determination method using a multidrug-resistance regulator protein as a framework.
Biochem.Biophys.Res.Commun., 518, 2019
|
|
6KO8
 
 | | Crystal structure of the Cholic acid bound RamR determined with XtaLAB Synergy | | Descriptor: | CHOLIC ACID, Putative regulatory protein, SULFATE ION | | Authors: | Matsumoto, T, Nakashima, R, Yamano, A, Nishino, K. | | Deposit date: | 2019-08-08 | | Release date: | 2019-10-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Development of a structure determination method using a multidrug-resistance regulator protein as a framework.
Biochem.Biophys.Res.Commun., 518, 2019
|
|
3ALN
 
 | | Crystal Structure of human non-phosphorylated MKK4 kinase domain complexed with AMP-PNP | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 4, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Matsumoto, T, Kinoshita, T, Kirii, Y, Yokota, K, Hamada, K, Tada, T. | | Deposit date: | 2010-08-04 | | Release date: | 2010-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of MKK4 kinase domain reveal that substrate peptide binds to an allosteric site and induces an auto-inhibition state
Biochem.Biophys.Res.Commun., 400, 2010
|
|
3ALO
 
 | | Crystal structure of human non-phosphorylated MKK4 kinase domain ternary complex with AMP-PNP and p38 peptide | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 4, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Matsumoto, T, Kinoshita, T, Kirii, Y, Yokota, K, Hamada, K, Tada, T. | | Deposit date: | 2010-08-04 | | Release date: | 2010-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of MKK4 kinase domain reveal that substrate peptide binds to an allosteric site and induces an auto-inhibition state
Biochem.Biophys.Res.Commun., 400, 2010
|
|
7EAP
 
 | | Crystal structure of IpeA-XXXG complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Matsuzawa, T, Watanabe, M, Nakamichi, Y, Akita, H, Yaoi, K. | | Deposit date: | 2021-03-08 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural basis for the catalytic mechanism of the glycoside hydrolase family 3 isoprimeverose-producing oligoxyloglucan hydrolase from Aspergillus oryzae.
Febs Lett., 596, 2022
|
|
3WL2
 
 | | Monoclinic Lysozyme at 0.96 A resolution | | Descriptor: | 1,2-ETHANEDIOL, Lysozyme C, NITRATE ION, ... | | Authors: | Matsumoto, T, Yamano, A, Hasegawa, T, Maeyama, M. | | Deposit date: | 2013-11-06 | | Release date: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Evaluation of Rigaku XtaLAB P200
To be Published
|
|
3APB
 
 | |
