2YVO
 
 | | Crystal structure of NDX2 in complex with MG2+ and AMP from thermus thermophilus HB8 | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, MutT/nudix family protein | | Authors: | Wakamatsu, T, Nakagawa, N, Kuramitsu, S, Yokoyama, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-13 | | Release date: | 2008-02-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural basis for different substrate specificities of two ADP-ribose pyrophosphatases from Thermus thermophilus HB8
J.Bacteriol., 190, 2008
|
|
6O97
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with propylamycin and bound to mRNA and A-, P-, and E-site tRNAs at 2.75A resolution | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2-{[3-O-(2,6-diamino-2,6-dideoxy-beta-L-idopyranosyl)-beta-D-ribofuranosyl]oxy}-3-hydroxyc yclohexyl 2-amino-2,4-dideoxy-4-propyl-alpha-D-glucopyranoside, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Matsushita, T, Sati, G.C, Kondasinghe, N, Pirrone, M.G, Kato, T, Waduge, P, Kumar, H.S, Sanchon, A.C, Dobosz-Bartoszek, M, Shcherbakov, D, Juhas, M, Hobbie, S.N, Schrepfer, T, Chow, C.S, Polikanov, Y.S, Schacht, J, Vasella, A, Bottger, E.C, Crich, D. | | Deposit date: | 2019-03-13 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Design, Multigram Synthesis, and in Vitro and in Vivo Evaluation of Propylamycin: A Semisynthetic 4,5-Deoxystreptamine Class Aminoglycoside for the Treatment of Drug-Resistant Enterobacteriaceae and Other Gram-Negative Pathogens.
J. Am. Chem. Soc., 141, 2019
|
|
6KO7
 
 | | Crystal structure of the Ethidium bound RamR determined with XtaLAB Synergy | | Descriptor: | ETHIDIUM, Putative regulatory protein, SULFATE ION | | Authors: | Matsumoto, T, Nakashima, R, Yamano, A, Nishino, K. | | Deposit date: | 2019-08-08 | | Release date: | 2019-10-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Development of a structure determination method using a multidrug-resistance regulator protein as a framework.
Biochem.Biophys.Res.Commun., 518, 2019
|
|
6KO9
 
 | | Crystal structure of the Gefitinib Intermediate 1 bound RamR determined with XtaLAB Synergy | | Descriptor: | 4-[(3-chloranyl-4-fluoranyl-phenyl)amino]-7-methoxy-quinazolin-6-ol, Putative regulatory protein, SULFATE ION | | Authors: | Matsumoto, T, Nakashima, R, Yamano, A, Nishino, K. | | Deposit date: | 2019-08-08 | | Release date: | 2019-10-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Development of a structure determination method using a multidrug-resistance regulator protein as a framework.
Biochem.Biophys.Res.Commun., 518, 2019
|
|
6KO8
 
 | | Crystal structure of the Cholic acid bound RamR determined with XtaLAB Synergy | | Descriptor: | CHOLIC ACID, Putative regulatory protein, SULFATE ION | | Authors: | Matsumoto, T, Nakashima, R, Yamano, A, Nishino, K. | | Deposit date: | 2019-08-08 | | Release date: | 2019-10-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Development of a structure determination method using a multidrug-resistance regulator protein as a framework.
Biochem.Biophys.Res.Commun., 518, 2019
|
|
8GN9
 
 | | SPFH domain of Pyrococcus horikoshii stomatin | | Descriptor: | SODIUM ION, Stomatin homolog PH1511 | | Authors: | Komatsu, T, Matsui, I, Yokoyama, H. | | Deposit date: | 2022-08-23 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and mutational studies suggest key residues to determine whether stomatin SPFH domains form dimers or trimers.
Biochem Biophys Rep, 32, 2022
|
|
6AL3
 
 | | Lys49 PLA2 BPII derived from the venom of Protobothrops flavoviridis. | | Descriptor: | Basic phospholipase A2 BP-II, SULFATE ION | | Authors: | Matsui, T, Kamata, S, Suzuki, A, Oda-Ueda, N, Ogawa, T, Tanaka, Y. | | Deposit date: | 2018-09-05 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | SDS-induced oligomerization of Lys49-phospholipase A2from snake venom.
Sci Rep, 9, 2019
|
|
1QVC
 
 | | CRYSTAL STRUCTURE ANALYSIS OF SINGLE STRANDED DNA BINDING PROTEIN (SSB) FROM E.COLI | | Descriptor: | SINGLE STRANDED DNA BINDING PROTEIN MONOMER | | Authors: | Matsumoto, T, Morimoto, Y, Shibata, N, Shimamoto, N, Tsukihara, T, Yasuoka, N. | | Deposit date: | 1999-07-07 | | Release date: | 2000-06-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Roles of functional loops and the C-terminal segment of a single-stranded DNA binding protein elucidated by X-Ray structure analysis.
J.Biochem.(Tokyo), 127, 2000
|
|
3I8R
 
 | | Crystal structure of the heme oxygenase from Corynebacterium diphtheriae (HmuO) in complex with heme binding ditiothreitol (DTT) | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, Heme oxygenase, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Matsui, T, Unno, M, Ikeda-Saito, M. | | Deposit date: | 2009-07-10 | | Release date: | 2010-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Dioxygen activation for the self-degradation of heme: reaction mechanism and regulation of heme oxygenase.
Inorg.Chem., 49, 2010
|
|
5ZN7
 
 | |
5WX5
 
 | |
5YE4
 
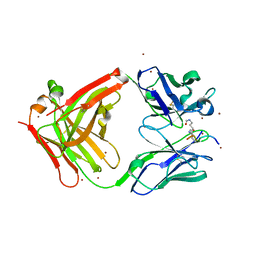 | | Crystal structure of the complex of di-acetylated histone H4 and 1A9D7 Fab fragment | | Descriptor: | 1A9D7 L chain, 1A9D7 VH CH1 chain, ZINC ION, ... | | Authors: | Matsuda, T, Ito, T, Wakamori, M, Umehara, T. | | Deposit date: | 2017-09-15 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | JQ1 affects BRD2-dependent and independent transcription regulation without disrupting H4-hyperacetylated chromatin states.
Epigenetics, 13, 2018
|
|
5WX4
 
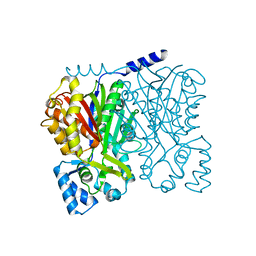 | |
5WX6
 
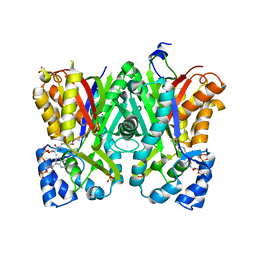 | | Alkyldiketide-CoA synthase W332Q mutant from Evodia rutaecarpa | | Descriptor: | Alkyldiketide-CoA synthase, COENZYME A, SULFATE ION | | Authors: | Matsui, T, Kodama, T, Tadakoshi, T, Morita, H. | | Deposit date: | 2017-01-06 | | Release date: | 2017-04-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | 2-Alkylquinolone alkaloid biosynthesis in the medicinal plant Evodia rutaecarpa involves collaboration of two novel type III polyketide synthases
J. Biol. Chem., 292, 2017
|
|
5WX3
 
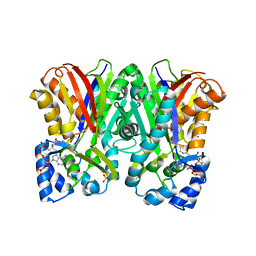 | | Alkyldiketide-CoA synthase from Evodia rutaecarpa | | Descriptor: | Alkyldiketide-CoA synthase, COENZYME A, SULFATE ION | | Authors: | Matsui, T, Kodama, T, Tadakoshi, T, Morita, H. | | Deposit date: | 2017-01-06 | | Release date: | 2017-04-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | 2-Alkylquinolone alkaloid biosynthesis in the medicinal plant Evodia rutaecarpa involves collaboration of two novel type III polyketide synthases
J. Biol. Chem., 292, 2017
|
|
5WX7
 
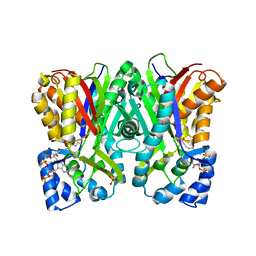 | | Alkyldiketide-CoA synthase W332G mutant from Evodia rutaecarpa | | Descriptor: | Alkyldiketide-CoA synthase, COENZYME A, SULFATE ION | | Authors: | Matsui, T, Kodama, T, Tadakoshi, T, Morita, H. | | Deposit date: | 2017-01-06 | | Release date: | 2017-04-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | 2-Alkylquinolone alkaloid biosynthesis in the medicinal plant Evodia rutaecarpa involves collaboration of two novel type III polyketide synthases
J. Biol. Chem., 292, 2017
|
|
5YE3
 
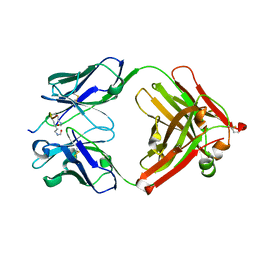 | | Crystal structure of the complex of di-acetylated histone H4 and 2A7D9 Fab fragment | | Descriptor: | 2A7D9 L chain, 2A7D9 VH CH1 chain, di-acetylated histone H4 | | Authors: | Matsuda, T, Ito, T, Wakamori, M, Umehara, T. | | Deposit date: | 2017-09-15 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | JQ1 affects BRD2-dependent and independent transcription regulation without disrupting H4-hyperacetylated chromatin states.
Epigenetics, 13, 2018
|
|
5XWD
 
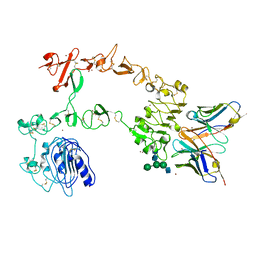 | | Crystal structure of the complex of 059-152-Fv and EGFR-ECD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Epidermal growth factor receptor, ... | | Authors: | Matsuda, T, Ito, T, Shirouzu, M. | | Deposit date: | 2017-06-29 | | Release date: | 2018-02-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.894 Å) | | Cite: | Cell-free synthesis of functional antibody fragments to provide a structural basis for antibody-antigen interaction
PLoS ONE, 13, 2018
|
|
5Z98
 
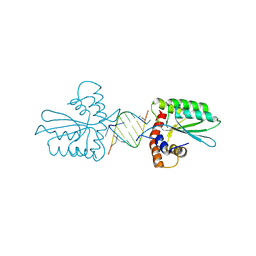 | | Crystal Structure of the Primate APOBEC3H Dimer mediated by RNA Duplex | | Descriptor: | Apolipoprotein B mRNA editing enzyme catalytic polypeptide-like protein 3H, RNA (5'-R(*AP*UP*AP*CP*CP*CP*GP*GP*CP*A)-3'), RNA (5'-R(P*CP*UP*GP*CP*CP*GP*GP*GP*UP*A)-3'), ... | | Authors: | Matsuoka, T, Nagae, T, Ode, H, Watanabe, N, Iwatani, Y. | | Deposit date: | 2018-02-02 | | Release date: | 2018-08-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of chimpanzee APOBEC3H dimerization stabilized by double-stranded RNA.
Nucleic Acids Res., 46, 2018
|
|
1EQQ
 
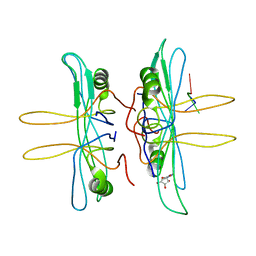 | | SINGLE STRANDED DNA BINDING PROTEIN AND SSDNA COMPLEX | | Descriptor: | 5'-R(*(5MU)P*(5MU)P*(5MU))-3', SINGLE STRANDED DNA BINDING PROTEIN | | Authors: | Matsumoto, T, Morimoto, Y, Shibata, N, Yasuoka, N, Shimamoto, N. | | Deposit date: | 2000-04-06 | | Release date: | 2003-09-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Roles of functional loops and the C-terminal segment of a single-stranded DNA binding protein elucidated by X-Ray structure analysis
J.Biochem.(Tokyo), 127, 2000
|
|
5B37
 
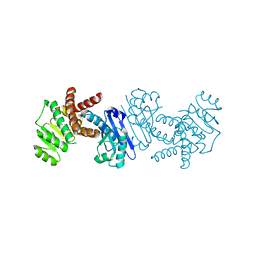 | | Crystal structure of L-tryptophan dehydrogenase from Nostoc punctiforme | | Descriptor: | Tryptophan dehydrogenase | | Authors: | Wakamatsu, T, Sakuraba, H, Kitamura, M, Hakumai, Y, Ohnishi, K, Ashiuchi, M, Ohshima, T. | | Deposit date: | 2016-02-11 | | Release date: | 2016-11-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Insights into l-Tryptophan Dehydrogenase from a Photoautotrophic Cyanobacterium, Nostoc punctiforme.
Appl. Environ. Microbiol., 83, 2017
|
|
3I9T
 
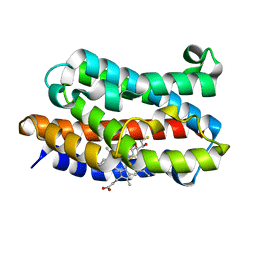 | | Crystal structure of the rat heme oxygenase (HO-1) in complex with heme binding dithiothreitol (DTT) | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, Heme oxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Matsui, T, Unno, M, Ikeda-Saito, M. | | Deposit date: | 2009-07-13 | | Release date: | 2010-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Dioxygen activation for the self-degradation of heme: reaction mechanism and regulation of heme oxygenase.
Inorg.Chem., 49, 2010
|
|
3I9U
 
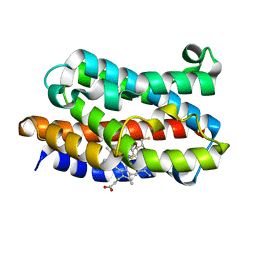 | | Crystal structure of the rat heme oxygenase (HO-1) in complex with heme binding dithioerythritol (DTE) | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, Heme oxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Matsui, T, Unno, M, Ikeda-Saito, M. | | Deposit date: | 2009-07-13 | | Release date: | 2010-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Dioxygen activation for the self-degradation of heme: reaction mechanism and regulation of heme oxygenase.
Inorg.Chem., 49, 2010
|
|
5GLK
 
 | | Crystal structure of CoXyl43, GH43 beta-xylosidase/alpha-arabinofuranosidase from a compost microbial metagenome, calcium-free form. | | Descriptor: | ACETATE ION, GLYCEROL, Glycoside hydrolase family 43, ... | | Authors: | Matsuzawa, T, Kishine, N, Fujimoto, Z, Yaoi, K. | | Deposit date: | 2016-07-12 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of metagenomic beta-xylosidase/ alpha-l-arabinofuranosidase activated by calcium.
J. Biochem., 162, 2017
|
|
5GLO
 
 | | Crystal structure of CoXyl43, GH43 beta-xylosidase/alpha-arabinofuranosidase from a compostmicrobial metagenome in complex with l-arabinose, calcium-free form | | Descriptor: | ACETATE ION, Glycoside hydrolase family 43, SODIUM ION, ... | | Authors: | Matsuzawa, T, Kishine, N, Fujimoto, Z, Yaoi, K. | | Deposit date: | 2016-07-12 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of metagenomic beta-xylosidase/ alpha-l-arabinofuranosidase activated by calcium.
J. Biochem., 162, 2017
|
|
