1TB3
 
 | | Crystal Structure Analysis of Recombinant Rat Kidney Long-chain Hydroxy Acid Oxidase | | Descriptor: | ACETIC ACID, FLAVIN MONONUCLEOTIDE, Hydroxyacid oxidase 3 | | Authors: | Cunane, L.M, Barton, J.D, Chen, Z.W, Le, K.H.D, Amar, D, Lederer, F, Mathews, F.S. | | Deposit date: | 2004-05-19 | | Release date: | 2005-02-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure Analysis of Recombinant Rat Kidney Long Chain Hydroxy Acid Oxidase.
Biochemistry, 44, 2005
|
|
1QCW
 
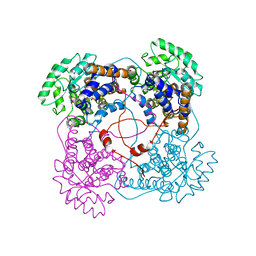 | | Flavocytochrome B2, ARG289LYS mutant | | Descriptor: | FLAVOCYTOCHROME B2, N-SULFO-FLAVIN MONONUCLEOTIDE | | Authors: | Mowat, C.G, Durley, R.C.E, Pike, A.D, Barton, J.D, Chen, Z.-W, Mathews, F.S, Lederer, F, Reid, G.A, Chapman, S.K. | | Deposit date: | 1999-05-07 | | Release date: | 1999-05-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Kinetic and crystallographic studies on the active site Arg289Lys mutant of flavocytochrome b2 (yeast L-lactate dehydrogenase)
Biochemistry, 39, 2000
|
|
1WVE
 
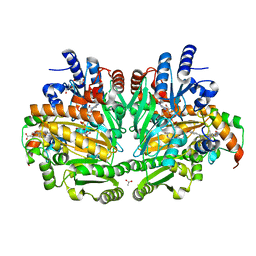 | | p-Cresol Methylhydroxylase: Alteration of the Structure of the Flavoprotein Subunit upon its Binding to the Cytochrome Subunit | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-cresol dehydrogenase [hydroxylating] cytochrome c subunit, 4-cresol dehydrogenase [hydroxylating] flavoprotein subunit, ... | | Authors: | Cunane, L.M, Chen, Z.-W, McIntire, W.S, Mathews, F.S. | | Deposit date: | 2004-12-15 | | Release date: | 2005-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | p-Cresol Methylhydroxylase: Alteration of the Structure of the Flavoprotein Subunit upon Its Binding to the Cytochrome Subunit
Biochemistry, 44, 2005
|
|
1WVF
 
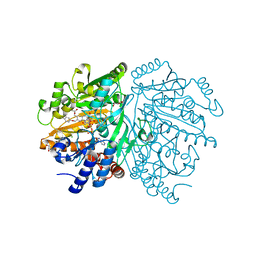 | | p-Cresol Methylhydroxylase: Alteration of the Structure of the Flavoprotein Subunit upon its Binding to the Cytochrome Subunit | | Descriptor: | 4-cresol dehydrogenase [hydroxylating] flavoprotein subunit, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Cunane, L.M, Chen, Z.-W, McIntire, W.S, Mathews, F.S. | | Deposit date: | 2004-12-15 | | Release date: | 2005-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | p-Cresol Methylhydroxylase: Alteration of the Structure of the Flavoprotein Subunit upon Its Binding to the Cytochrome Subunit
Biochemistry, 44, 2005
|
|
1YIQ
 
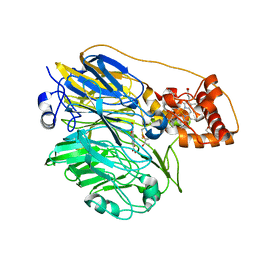 | | Molecular cloning and structural analysis of quinohemoprotein alcohol dehydrogenase ADHIIG from Pseudomonas putida HK5. Compariison to the other quinohemoprotein alcohol dehydrogenase ADHIIB found in the same microorganism. | | Descriptor: | CALCIUM ION, HEME C, PYRROLOQUINOLINE QUINONE, ... | | Authors: | Toyama, H, Chen, Z.W, Fukumoto, M, Adachi, O, Matsushita, K, Mathews, F.S. | | Deposit date: | 2005-01-12 | | Release date: | 2005-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular cloning and structural analysis of quinohemoprotein alcohol dehydrogenase ADH-IIG from Pseudomonas putida HK5
J.Mol.Biol., 352, 2005
|
|
2A89
 
 | | Monomeric Sarcosine Oxidase: Structure of a covalently flavinylated amine oxidizing enzyme | | Descriptor: | (N5,C4A)-(ALPHA-HYDROXY-PROPANO)-3,4,4A,5-TETRAHYDRO-FLAVIN-ADENINE DINUCLEOTIDE, CHLORIDE ION, Monomeric sarcosine oxidase, ... | | Authors: | Chen, Z.-W, Zhao, G, Martinovic, S, Jorns, M.S, Mathews, F.S. | | Deposit date: | 2005-07-07 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the sodium borohydride-reduced N-(cyclopropyl)glycine adduct of the flavoenzyme monomeric sarcosine oxidase.
Biochemistry, 44, 2005
|
|
2IDQ
 
 | | Structure of M98A mutant of amicyanin, Cu(II) | | Descriptor: | Amicyanin, COPPER (II) ION, PHOSPHATE ION | | Authors: | Carrell, C.J, Ma, J.K, Antholine, W, Hosler, J.P, Mathews, F.S, Davidson, V.L. | | Deposit date: | 2006-09-15 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Generation of Novel Copper Sites by Mutation of the Axial Ligand of Amicyanin. Atomic Resolution Structures and Spectroscopic Properties
Biochemistry, 46, 2007
|
|
1BXA
 
 | | AMICYANIN REDUCED, PH 4.4, 1.3 ANGSTROMS | | Descriptor: | COPPER (I) ION, PROTEIN (AMICYANIN) | | Authors: | Cunane, L.M, Chen, Z.W, Durley, R.C.E, Mathews, F.S. | | Deposit date: | 1998-10-01 | | Release date: | 1998-10-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular basis for interprotein complex-dependent effects on the redox properties of amicyanin.
Biochemistry, 37, 1998
|
|
2IAA
 
 | | Crystal Structure of an Electron Transfer Complex Between Aromatic Amine Dephydrogenase and Azurin from Alcaligenes Faecalis (Form 2) | | Descriptor: | Aromatic Amine Dehydrogenase, Azurin, COPPER (II) ION | | Authors: | Sukumar, N, Chen, Z, Leys, D, Scrutton, N.S, Ferrati, D, Merli, A, Rossi, G.L, Bellamy, H.D, Chistoserdov, A, Davidson, V.L, Mathews, F.S. | | Deposit date: | 2006-09-07 | | Release date: | 2006-11-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of an Electron Transfer Complex between Aromatic Amine Dehydrogenase and Azurin from Alcaligenes faecalis.
Biochemistry, 45, 2006
|
|
2IDS
 
 | | Structure of M98A mutant of amicyanin, Cu(I) | | Descriptor: | Amicyanin, COPPER (I) ION | | Authors: | Carrell, C.J, Ma, J.K, Antholine, W, Hosler, J.P, Mathews, F.S, Davidson, V.L. | | Deposit date: | 2006-09-15 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Generation of Novel Copper Sites by Mutation of the Axial Ligand of Amicyanin. Atomic Resolution Structures and Spectroscopic Properties
Biochemistry, 46, 2007
|
|
2IDU
 
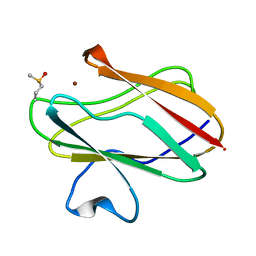 | | Structure of M98Q mutant of amicyanin, Cu(I) | | Descriptor: | Amicyanin, COPPER (I) ION | | Authors: | Carrell, C.J, Ma, J.K, Antholine, W, Hosler, J.P, Mathews, F.S, Davidson, V.L. | | Deposit date: | 2006-09-15 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Generation of Novel Copper Sites by Mutation of the Axial Ligand of Amicyanin. Atomic Resolution Structures and Spectroscopic Properties
Biochemistry, 46, 2007
|
|
1ASH
 
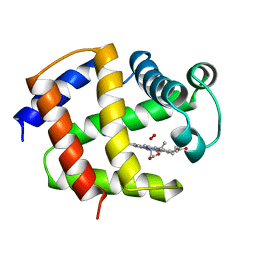 | | THE STRUCTURE OF ASCARIS HEMOGLOBIN DOMAIN I AT 2.2 ANGSTROMS RESOLUTION: MOLECULAR FEATURES OF OXYGEN AVIDITY | | Descriptor: | HEMOGLOBIN (OXY), OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yang, J, Mathews, F.S, Kloek, A.P, Goldberg, D.E. | | Deposit date: | 1995-01-06 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structure of Ascaris hemoglobin domain I at 2.2 A resolution: molecular features of oxygen avidity.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
2IDT
 
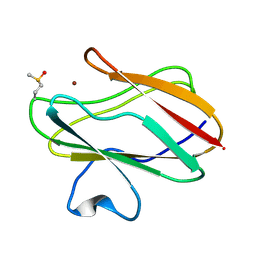 | | Structure of M98Q mutant of amicyanin, Cu(II) | | Descriptor: | Amicyanin, COPPER (II) ION | | Authors: | Carrell, C.J, Ma, J.K, Antholine, W, Hosler, J.P, Mathews, F.S, Davidson, V.L. | | Deposit date: | 2006-09-15 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Generation of Novel Copper Sites by Mutation of the Axial Ligand of Amicyanin. Atomic Resolution Structures and Spectroscopic Properties
Biochemistry, 46, 2007
|
|
2H47
 
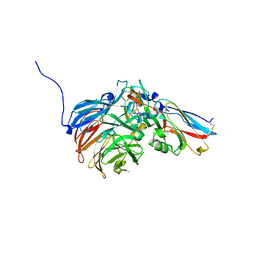 | | Crystal Structure of an Electron Transfer Complex Between Aromatic Amine Dephydrogenase and Azurin from Alcaligenes Faecalis (Form 1) | | Descriptor: | Aromatic Amine Dehydrogenase, Azurin, COPPER (II) ION | | Authors: | Sukumar, N, Chen, Z, Leys, D, Scrutton, N.S, Ferrati, D, Merli, A, Rossi, G.L, Bellamy, H.D, Chistoserdov, A, Davidson, V.L, Mathews, F.S. | | Deposit date: | 2006-05-23 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of an Electron Transfer Complex between Aromatic Amine Dehydrogenase and Azurin from Alcaligenes faecalis.
Biochemistry, 45, 2006
|
|
2H3X
 
 | | Crystal Structure of an Electron Transfer Complex Between Aromatic Amine Dehydrogenase and Azurin from Alcaligenes Faecalis (Form 3) | | Descriptor: | Aromatic Amine Dehydrogenase, Azurin, COPPER (II) ION | | Authors: | Sukumar, N, Chen, Z, Leys, D, Scrutton, N.S, Ferrati, D, Merli, A, Rossi, G.L, Bellamy, H.D, Chistoserdov, A, Davidson, V.L, Mathews, F.S. | | Deposit date: | 2006-05-23 | | Release date: | 2006-11-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of an Electron Transfer Complex between Aromatic Amine Dehydrogenase and Azurin from Alcaligenes faecalis.
Biochemistry, 45, 2006
|
|
1DJN
 
 | | STRUCTURAL AND BIOCHEMICAL CHARACTERIZATION OF RECOMBINANT WILD TYPE TRIMETHYLAMINE DEHYDROGENASE FROM METHYLOPHILUS METHYLOTROPHUS (SP. W3A1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Trickey, P, Basran, J, Lian, L.-Y, Chen, Z.-W, Barton, J.D, Sutcliffe, M.J, Scrutton, N.S, Mathews, F.S. | | Deposit date: | 1999-12-03 | | Release date: | 1999-12-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and biochemical characterization of recombinant wild type and a C30A mutant of trimethylamine dehydrogenase from methylophilus methylotrophus (sp. W(3)A(1)).
Biochemistry, 39, 2000
|
|
1DII
 
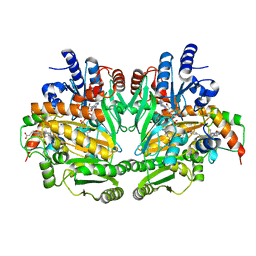 | | CRYSTAL STRUCTURE OF P-CRESOL METHYLHYDROXYLASE AT 2.5 A RESOLUTION | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, HEME C, ... | | Authors: | Cunane, L.M, Chen, Z.W, Shamala, N, Mathews, F.S, Cronin, C.N, McIntire, W.S. | | Deposit date: | 1999-11-29 | | Release date: | 1999-12-08 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the flavocytochrome p-cresol methylhydroxylase and its enzyme-substrate complex: gated substrate entry and proton relays support the proposed catalytic mechanism.
J.Mol.Biol., 295, 2000
|
|
1AAC
 
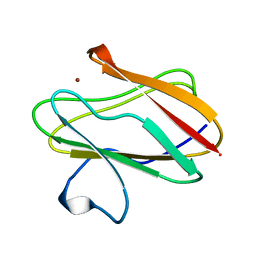 | | AMICYANIN OXIDIZED, 1.31 ANGSTROMS | | Descriptor: | AMICYANIN, COPPER (II) ION | | Authors: | Cunane, L.M, Chen, Z.-W, Durley, R.C.E, Mathews, F.S. | | Deposit date: | 1995-09-07 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | X-ray structure of the cupredoxin amicyanin, from Paracoccus denitrificans, refined at 1.31 A resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1DIQ
 
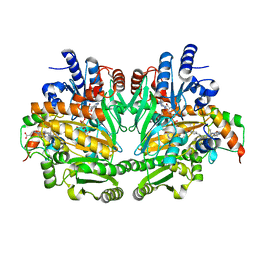 | | CRYSTAL STRUCTURE OF P-CRESOL METHYLHYDROXYLASE WITH SUBSTRATE BOUND | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, HEME C, ... | | Authors: | Cunane, L.M, Chen, Z.W, Shamala, N, Mathews, F.S, Cronin, C.S, McIntire, W.S. | | Deposit date: | 1999-11-29 | | Release date: | 1999-12-08 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structures of the flavocytochrome p-cresol methylhydroxylase and its enzyme-substrate complex: gated substrate entry and proton relays support the proposed catalytic mechanism.
J.Mol.Biol., 295, 2000
|
|
1DJQ
 
 | | STRUCTURAL AND BIOCHEMICAL CHARACTERIZATION OF RECOMBINANT C30A MUTANT OF TRIMETHYLAMINE DEHYDROGENASE FROM METHYLOPHILUS METHYLOTROPHUS (SP. W3A1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Trickey, P, Basran, J, Lian, L.-Y, Chen, Z.-W, Barton, J.D, Sutcliffe, M.J, Scrutton, N.S, Mathews, F.S. | | Deposit date: | 1999-12-03 | | Release date: | 1999-12-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and biochemical characterization of recombinant wild type and a C30A mutant of trimethylamine dehydrogenase from methylophilus methylotrophus (sp. W(3)A(1)).
Biochemistry, 39, 2000
|
|
2HWL
 
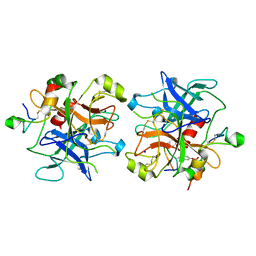 | | Crystal structure of thrombin in complex with fibrinogen gamma' peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fibrinogen gamma' peptide, Prothrombin, ... | | Authors: | Pineda, A.O, Chen, Z.W, Marino, F, Mathews, F.S, Mosesson, M.W, Di Cera, E. | | Deposit date: | 2006-08-01 | | Release date: | 2006-09-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of thrombin in complex with fibrinogen gamma' peptide.
Biophys.Chem., 125, 2007
|
|
1EKM
 
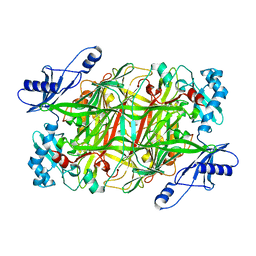 | | CRYSTAL STRUCTURE AT 2.5 A RESOLUTION OF ZINC-SUBSTITUTED COPPER AMINE OXIDASE OF HANSENULA POLYMORPHA EXPRESSED IN ESCHERICHIA COLI | | Descriptor: | COPPER AMINE OXIDASE, ZINC ION | | Authors: | Chen, Z, Schwartz, B, Williams, N.K, Li, R, Klinman, J.P, Mathews, F.S. | | Deposit date: | 2000-03-09 | | Release date: | 2000-08-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure at 2.5 A resolution of zinc-substituted copper amine oxidase of Hansenula polymorpha expressed in Escherichia coli.
Biochemistry, 39, 2000
|
|
1ELI
 
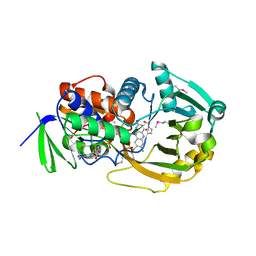 | | COMPLEX OF MONOMERIC SARCOSINE OXIDASE WITH THE INHIBITOR PYRROLE-2-CARBOXYLATE | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION, ... | | Authors: | Wagner, M.A, Trickey, P, Chen, Z.-W, Mathews, F.S, Jorns, M.S. | | Deposit date: | 2000-03-13 | | Release date: | 2000-12-06 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Monomeric sarcosine oxidase: 1. Flavin reactivity and active site binding determinants.
Biochemistry, 39, 2000
|
|
1EL7
 
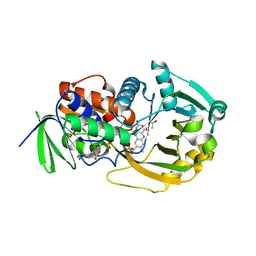 | | COMPLEX OF MONOMERIC SARCOSINE OXIDASE WITH THE INHIBITOR [METHYTELLURO]ACETATE | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION, ... | | Authors: | Wagner, M.A, Trickey, P, Chen, Z.-W, Mathews, F.S, Jorns, M.S. | | Deposit date: | 2000-03-13 | | Release date: | 2000-12-06 | | Last modified: | 2023-02-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Monomeric sarcosine oxidase: 1. Flavin reactivity and active site binding determinants.
Biochemistry, 39, 2000
|
|
1PBY
 
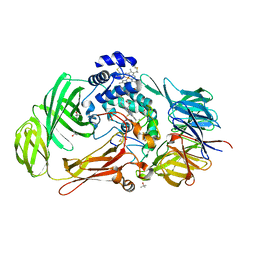 | | Structure of the Phenylhydrazine Adduct of the Quinohemoprotein Amine Dehydrogenase from Paracoccus denitrificans at 1.7 A Resolution | | Descriptor: | HEME C, TERTIARY-BUTYL ALCOHOL, quinohemoprotein amine dehydrogenase 40 kDa subunit, ... | | Authors: | Datta, S, Ikeda, T, Kano, K, Mathews, F.S. | | Deposit date: | 2003-05-15 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the phenylhydrazine adduct of the quinohemoprotein amine dehydrogenase from Paracoccus denitrificans at 1.7 A resolution.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
