1V58
 
 | | Crystal Structure Of the Reduced Protein Disulfide Bond Isomerase DsbG | | Descriptor: | SULFATE ION, Thiol:disulfide interchange protein dsbG | | Authors: | Heras, B, Edeling, M.A, Schirra, H.J, Raina, S, Martin, J.L. | | Deposit date: | 2003-11-21 | | Release date: | 2004-06-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the DsbG disulfide isomerase reveal an unstable disulfide
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1YZ3
 
 | | Structure of human pnmt complexed with cofactor product adohcy and inhibitor SK&F 64139 | | Descriptor: | 7,8-DICHLORO-1,2,3,4-TETRAHYDROISOQUINOLINE, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Wu, Q, Gee, C.L, Lin, F, Martin, J.L, Grunewald, G.L, McLeish, M.J. | | Deposit date: | 2005-02-27 | | Release date: | 2006-02-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural, mutagenic, and kinetic analysis of the binding of substrates and inhibitors of human phenylethanolamine N-methyltransferase
J.Med.Chem., 48, 2005
|
|
5VYO
 
 | |
1V57
 
 | | Crystal Structure of the Disulfide Bond Isomerase DsbG | | Descriptor: | SULFATE ION, Thiol:disulfide interchange protein dsbG | | Authors: | Heras, B, Edeling, M.A, Schirra, H.J, Raina, S, Martin, J.L. | | Deposit date: | 2003-11-21 | | Release date: | 2004-06-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the DsbG disulfide isomerase reveal an unstable disulfide
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1J1A
 
 | | PANCREATIC SECRETORY PHOSPHOLIPASE A2 (IIa) WITH ANTI-INFLAMMATORY ACTIVITY | | Descriptor: | (S)-5-(4-BENZYLOXY-PHENYL)-4-(7-PHENYL-HEPTANOYLAMINO)-PENTANOIC ACID, CALCIUM ION, Phospholipase A2 | | Authors: | Hansford, K.A, Reid, R.C, Clark, C.I, Tyndall, J.D.A, Whitehouse, M.W, Guthrie, T, McGeary, R.P, Schafer, K, Martin, J.L, Fairlie, D.P. | | Deposit date: | 2002-12-03 | | Release date: | 2003-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | D-Tyrosine as a Chiral Precusor to Potent Inhibitors of Human Nonpancreatic Secretory Phospholipase A2 (IIa) with Antiinflammatory Activity
Chembiochem, 4, 2003
|
|
1PEN
 
 | | ALPHA-CONOTOXIN PNI1 | | Descriptor: | ALPHA-CONOTOXIN PNIA | | Authors: | Hu, S.-H, Gehrmann, J, Guddat, L.W, Alewood, P.F, Craik, D.J, Martin, J.L. | | Deposit date: | 1996-01-29 | | Release date: | 1997-04-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The 1.1 A crystal structure of the neuronal acetylcholine receptor antagonist, alpha-conotoxin PnIA from Conus pennaceus.
Structure, 4, 1996
|
|
1N7I
 
 | | The structure of Phenylethanolamine N-methyltransferase in complex with S-adenosylhomocysteine and the inhibitor LY134046 | | Descriptor: | 8,9-DICHLORO-2,3,4,5-TETRAHYDRO-1H-BENZO[C]AZEPINE, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | McMillan, F.M, Archbold, J, McLeish, M.J, Caine, J.M, Criscione, K.R, Grunewald, G.L, Martin, J.L. | | Deposit date: | 2002-11-15 | | Release date: | 2003-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular recognition of sub-micromolar inhibitors by the epinephrine-synthesizing enzyme phenylethanolamine N-methyltransferase.
J.Med.Chem., 47, 2004
|
|
1N7J
 
 | | The structure of Phenylethanolamine N-methyltransferase in complex with S-adenosylhomocysteine and an iodinated inhibitor | | Descriptor: | 7-IODO-1,2,3,4-TETRAHYDRO-ISOQUINOLINE, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | McMillan, F.M, Archbold, J, McLeish, M.J, Caine, J.M, Criscione, K.R, Grunewald, G.L, Martin, J.L. | | Deposit date: | 2002-11-15 | | Release date: | 2003-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular recognition of sub-micromolar inhibitors by the epinephrine-synthesizing enzyme phenylethanolamine N-methyltransferase.
J.Med.Chem., 47, 2004
|
|
1G9S
 
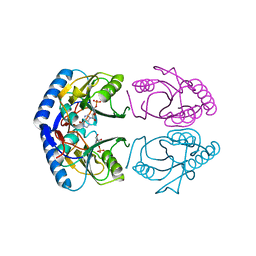 | | CRYSTAL STRUCTURE OF A COMPLEX BETWEEN E.COLI HPRT AND IMP | | Descriptor: | ANY 5'-MONOPHOSPHATE NUCLEOTIDE, HYPOXANTHINE PHOSPHORIBOSYLTRANSFERASE, INOSINIC ACID | | Authors: | Guddat, L.W, Vos, S, Martin, J.L, Keough, D.T, de Jersey, J. | | Deposit date: | 2000-11-27 | | Release date: | 2002-08-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of free, IMP-, and GMP-bound Escherichia coli hypoxanthine phosphoribosyltransferase.
Protein Sci., 11, 2002
|
|
1G9T
 
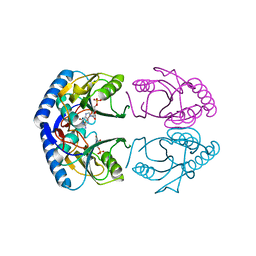 | | CRYSTAL STRUCTURE OF E.COLI HPRT-GMP COMPLEX | | Descriptor: | ANY 5'-MONOPHOSPHATE NUCLEOTIDE, GUANOSINE-5'-MONOPHOSPHATE, HYPOXANTHINE PHOSPHORIBOSYLTRANSFERASE | | Authors: | Guddat, L.W, Vos, S, Martin, J.L, keough, D.T, de Jersey, J. | | Deposit date: | 2000-11-28 | | Release date: | 2002-08-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of free, IMP-, and GMP-bound Escherichia coli hypoxanthine phosphoribosyltransferase.
Protein Sci., 11, 2002
|
|
3KR2
 
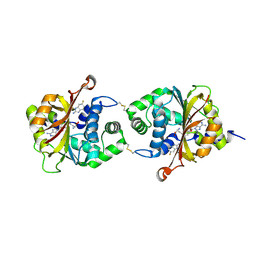 | |
1GRV
 
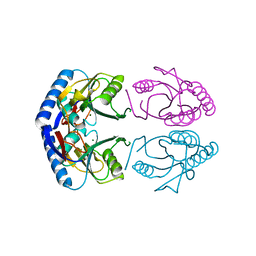 | | Hypoxanthine Phosphoribosyltransferase from E. coli | | Descriptor: | HYPOXANTHINE PHOSPHORIBOSYLTRANSFERASE, MAGNESIUM ION | | Authors: | Guddat, L.W, Vos, S, Martin, J.L, Keough, D.T, De Jersey, J. | | Deposit date: | 2001-12-17 | | Release date: | 2002-12-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structures of Free, Imp-, and Gmp- Bound Escherichia Coli Hypoxanthine Phosphoribosyltransferase
Protein Sci., 11, 2002
|
|
3KQY
 
 | |
3KQO
 
 | | Crystal Structure of hPNMT in Complex AdoHcy and 6-Chloropurine | | Descriptor: | 6-chloro-9H-purine, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Drinkwater, N, Martin, J.L. | | Deposit date: | 2009-11-17 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Fragment-based screening by X-ray crystallography, MS and isothermal titration calorimetry to identify PNMT (phenylethanolamine N-methyltransferase) inhibitors.
Biochem.J., 431, 2010
|
|
3KR1
 
 | |
1LS6
 
 | | Human SULT1A1 complexed with PAP and p-Nitrophenol | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, P-NITROPHENOL, aryl sulfotransferase | | Authors: | Gamage, N.U, Barnett, A.C, Tresillian, M, Latham, C.F, Liyou, N.E, McManus, M.E, Martin, J.L. | | Deposit date: | 2002-05-17 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a human carcinogen-converting enzyme, SULT1A1. Structural and kinetic implications of substrate inhibition.
J.Biol.Chem., 278, 2003
|
|
1NOT
 
 | | THE 1.2 ANGSTROM STRUCTURE OF G1 ALPHA CONOTOXIN | | Descriptor: | GI ALPHA CONOTOXIN | | Authors: | Guddat, L.W, Shan, L, Martin, J.L, Edmundson, A.B, Gray, W.R. | | Deposit date: | 1996-05-02 | | Release date: | 1996-12-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Three-dimensional structure of the alpha-conotoxin GI at 1.2 A resolution
Biochemistry, 35, 1996
|
|
3PUJ
 
 | | Crystal structure of the MUNC18-1 and SYNTAXIN4 N-Peptide complex | | Descriptor: | Syntaxin-4 N-terminal peptide, Syntaxin-binding protein 1 | | Authors: | Hu, S.-H, Christie, M.P, Saez, N.J, Latham, C.F, Jarrott, R, Lua, L.H.L, Collins, B.M, Martin, J.L. | | Deposit date: | 2010-12-05 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.313 Å) | | Cite: | Possible roles for Munc18-1 domain 3a and Syntaxin1 N-peptide and C-terminal anchor in SNARE complex formation
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3KQM
 
 | | Crystal Structure of hPNMT in Complex AdoHcy and 4-Bromo-1H-imidazole | | Descriptor: | 4-bromo-1H-imidazole, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Drinkwater, N, Martin, J.L. | | Deposit date: | 2009-11-17 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Fragment-based screening by X-ray crystallography, MS and isothermal titration calorimetry to identify PNMT (phenylethanolamine N-methyltransferase) inhibitors.
Biochem.J., 431, 2010
|
|
3KQS
 
 | | Crystal Structure of hPNMT in Complex AdoHcy and 2-Aminobenzimidazole | | Descriptor: | 1H-benzimidazol-2-amine, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Drinkwater, N, Martin, J.L. | | Deposit date: | 2009-11-17 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Fragment-based screening by X-ray crystallography, MS and isothermal titration calorimetry to identify PNMT (phenylethanolamine N-methyltransferase) inhibitors.
Biochem.J., 431, 2010
|
|
3KPV
 
 | | Crystal Structure of hPNMT in Complex AdoHcy and Adenine | | Descriptor: | ADENINE, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Drinkwater, N, Martin, J.L. | | Deposit date: | 2009-11-17 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Fragment-based screening by X-ray crystallography, MS and isothermal titration calorimetry to identify PNMT (phenylethanolamine N-methyltransferase) inhibitors.
Biochem.J., 431, 2010
|
|
3KPJ
 
 | | Crystal Structure of hPNMT in Complex AdoHcy and Bound Phosphate | | Descriptor: | PHOSPHATE ION, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Drinkwater, N, Martin, J.L. | | Deposit date: | 2009-11-16 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fragment-based screening by X-ray crystallography, MS and isothermal titration calorimetry to identify PNMT (phenylethanolamine N-methyltransferase) inhibitors.
Biochem.J., 431, 2010
|
|
3KQQ
 
 | | Crystal Structure of hPNMT in Complex AdoHcy and 2-Hydroxynicotinic acid | | Descriptor: | 2-oxo-1,2-dihydropyridine-3-carboxylic acid, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Drinkwater, N, Martin, J.L. | | Deposit date: | 2009-11-17 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fragment-based screening by X-ray crystallography, MS and isothermal titration calorimetry to identify PNMT (phenylethanolamine N-methyltransferase) inhibitors.
Biochem.J., 431, 2010
|
|
3KQW
 
 | | Crystal Structure of hPNMT in Complex AdoHcy and 5-Chlorobenzimidazole | | Descriptor: | 5-chloro-1H-benzimidazole, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Drinkwater, N, Martin, J.L. | | Deposit date: | 2009-11-17 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.486 Å) | | Cite: | Fragment-based screening by X-ray crystallography, MS and isothermal titration calorimetry to identify PNMT (phenylethanolamine N-methyltransferase) inhibitors.
Biochem.J., 431, 2010
|
|
3KPY
 
 | | Crystal Structure of hPNMT in Complex AdoHcy and 6-Chlorooxindole | | Descriptor: | 6-chloro-1,3-dihydro-2H-indol-2-one, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Drinkwater, N, Martin, J.L. | | Deposit date: | 2009-11-17 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Fragment-based screening by X-ray crystallography, MS and isothermal titration calorimetry to identify PNMT (phenylethanolamine N-methyltransferase) inhibitors.
Biochem.J., 431, 2010
|
|
