7STX
 
 | |
7SNG
 
 | | structure of G6PD-WT tetramer | | Descriptor: | Glucose-6-phosphate 1-dehydrogenase | | Authors: | Wei, X, Marmorstein, R. | | Deposit date: | 2021-10-28 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Allosteric role of a structural NADP + molecule in glucose-6-phosphate dehydrogenase activity.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7SNF
 
 | | Structure of G6PD-WT dimer | | Descriptor: | Glucose-6-phosphate 1-dehydrogenase | | Authors: | Wei, X, Marmorstein, R. | | Deposit date: | 2021-10-28 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Allosteric role of a structural NADP + molecule in glucose-6-phosphate dehydrogenase activity.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7SNH
 
 | | Structure of G6PD-D200N tetramer bound to NADP+ | | Descriptor: | Glucose-6-phosphate 1-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Wei, X, Marmorstein, R. | | Deposit date: | 2021-10-28 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Allosteric role of a structural NADP + molecule in glucose-6-phosphate dehydrogenase activity.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5J8C
 
 | | Human MOF C316S, E350Q crystal structure | | Descriptor: | CHLORIDE ION, Histone acetyltransferase KAT8, SODIUM ION, ... | | Authors: | McCullough, C.E, Marmorstein, R. | | Deposit date: | 2016-04-07 | | Release date: | 2016-07-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural and Functional Role of Acetyltransferase hMOF K274 Autoacetylation.
J.Biol.Chem., 291, 2016
|
|
5J8F
 
 | | Human MOF K274P crystal structure | | Descriptor: | CHLORIDE ION, Histone acetyltransferase KAT8, ZINC ION | | Authors: | McCullough, C.E, Marmorstein, R. | | Deposit date: | 2016-04-07 | | Release date: | 2016-07-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Functional Role of Acetyltransferase hMOF K274 Autoacetylation.
J.Biol.Chem., 291, 2016
|
|
1FY7
 
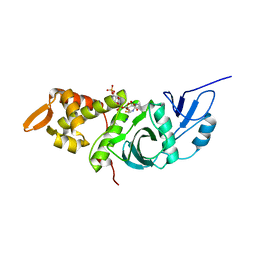 | | CRYSTAL STRUCTURE OF YEAST ESA1 HISTONE ACETYLTRANSFERASE DOMAIN COMPLEXED WITH COENZYME A | | Descriptor: | COENZYME A, ESA1 HISTONE ACETYLTRANSFERASE, SODIUM ION | | Authors: | Yan, Y, Barlev, N.A, Haley, R.H, Berger, S.L, Marmorstein, R. | | Deposit date: | 2000-09-28 | | Release date: | 2000-11-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of yeast Esa1 suggests a unified mechanism for catalysis and substrate binding by histone acetyltransferases.
Mol.Cell, 6, 2000
|
|
4RLO
 
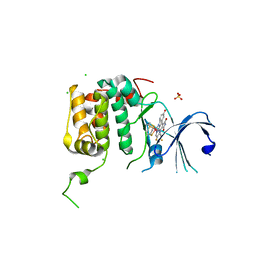 | | Human p70s6k1 with ruthenium-based inhibitor EM5 | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Domsic, J.F, Barber-Rotenberg, J, Salami, J, Qin, J, Marmorstein, R. | | Deposit date: | 2014-10-17 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.527 Å) | | Cite: | Development of Organometallic S6K1 Inhibitors.
J.Med.Chem., 58, 2015
|
|
4U9W
 
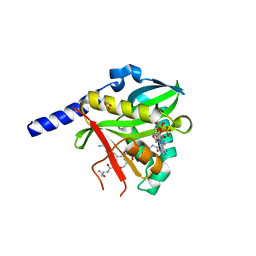 | | Crystal Structure of NatD bound to H4/H2A peptide and CoA | | Descriptor: | COENZYME A, GLYCEROL, Histone H4/H2A N-terminus, ... | | Authors: | Magin, R.S, Liszczak, G.P, Marmorstein, R. | | Deposit date: | 2014-08-06 | | Release date: | 2015-01-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | The Molecular Basis for Histone H4- and H2A-Specific Amino-Terminal Acetylation by NatD.
Structure, 23, 2015
|
|
3TOB
 
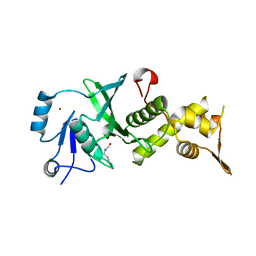 | |
7KD7
 
 | |
4PO2
 
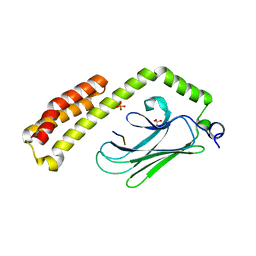 | | Crystal Structure of the Stress-Inducible Human Heat Shock Protein HSP70 Substrate-Binding Domain in Complex with Peptide Substrate | | Descriptor: | HSP70 substrate peptide, Heat shock 70 kDa protein 1A/1B, PHOSPHATE ION, ... | | Authors: | Zhang, P, Leu, J.I, Murphy, M.E, George, D.L, Marmorstein, R. | | Deposit date: | 2014-02-24 | | Release date: | 2014-08-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the stress-inducible human heat shock protein 70 substrate-binding domain in complex with Peptide substrate.
Plos One, 9, 2014
|
|
4RLP
 
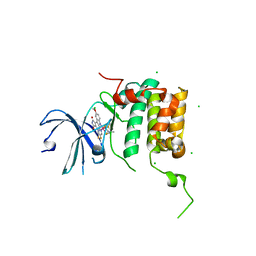 | | Human p70s6k1 with ruthenium-based inhibitor FL772 | | Descriptor: | CHLORIDE ION, [(amino-kappaN)methanethiolato](3-fluoro-9-methoxypyrido[2,3-a]pyrrolo[3,4-c]carbazole-5,7(6H,12H)-dionato-kappa~2~N,N')(N-methyl-1,4,7-trithiecan-9-amine-kappa~3~S~1~,S~4~,S~7~)ruthenium, p70S6K1 | | Authors: | Domsic, J.F, Barber-Rotenberg, J, Salami, J, Qin, J, Marmorstein, R. | | Deposit date: | 2014-10-17 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Development of Organometallic S6K1 Inhibitors.
J.Med.Chem., 58, 2015
|
|
4PZT
 
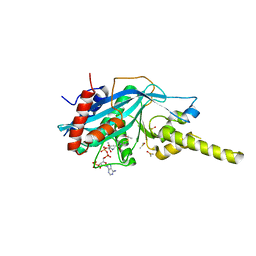 | | Crystal structure of p300 histone acetyltransferase domain in complex with an inhibitor, Acetonyl-Coenzyme A | | Descriptor: | DIMETHYL SULFOXIDE, Histone acetyltransferase p300, [(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-4-HYDROXY-3-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL (3R)-3-HYDROXY-2,2-DIMETHYL-4-OXO-4-{[3-OXO-3-({2-[(2-OXOPROPYL)THIO]ETHYL}AMINO)PROPYL]AMINO}BUTYL DIHYDROGEN DIPHOSPHATE | | Authors: | Maksimoska, J, Marmorstein, R. | | Deposit date: | 2014-03-31 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the p300 Histone Acetyltransferase Bound to Acetyl-Coenzyme A and Its Analogues.
Biochemistry, 53, 2014
|
|
1CS3
 
 | |
4U9V
 
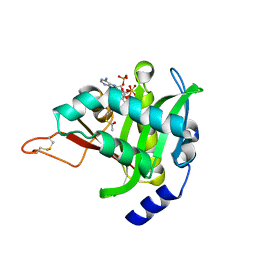 | |
4UA3
 
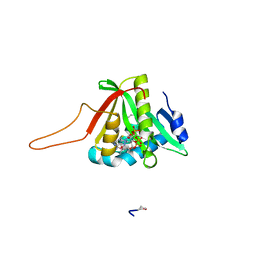 | |
4PZR
 
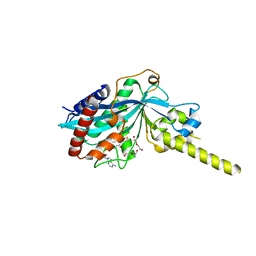 | |
4PZS
 
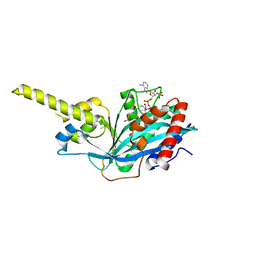 | |
3Q4C
 
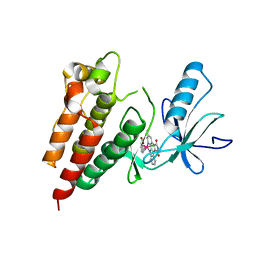 | | Crystal Structure of Wild Type BRAF kinase domain in complex with organometallic inhibitor CNS292 | | Descriptor: | Serine/threonine-protein kinase B-raf, [(1,2,3,4,5,6-eta)-(1S,2R,3R,4R,5S,6S)-1-carboxycyclohexane-1,2,3,4,5,6-hexayl](chloro)(3-methyl-5,7-dioxo-6,7-dihydro-5H-pyrido[2,3-a]pyrrolo[3,4-c]carbazol-12-ide-kappa~2~N~1~,N~12~)ruthenium(1+) | | Authors: | Xie, P, Streu, C, Qin, J, Pregman, H, Pagano, N, Meggers, E, Marmorstein, R. | | Deposit date: | 2010-12-23 | | Release date: | 2011-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The crystal structure of BRAF in complex with an organoruthenium inhibitor reveals a mechanism for inhibition of an active form of BRAF kinase.
Biochemistry, 48, 2009
|
|
7KPU
 
 | |
1DUX
 
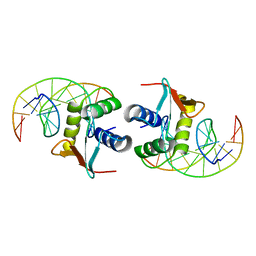 | | ELK-1/DNA STRUCTURE REVEALS HOW RESIDUES DISTAL FROM DNA-BINDING SURFACE AFFECT DNA-RECOGNITION | | Descriptor: | DNA (5'-D(*AP*CP*AP*CP*TP*TP*CP*CP*GP*GP*TP*CP*A)-3'), DNA (5'-D(*TP*GP*AP*CP*CP*GP*GP*AP*AP*GP*TP*GP*T)-3'), ETS-DOMAIN PROTEIN ELK-1 | | Authors: | Mo, Y, Vaessen, B, Johnston, K, Marmorstein, R. | | Deposit date: | 2000-01-19 | | Release date: | 2000-04-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the elk-1-DNA complex reveals how DNA-distal residues affect ETS domain recognition of DNA.
Nat.Struct.Biol., 7, 2000
|
|
3TO7
 
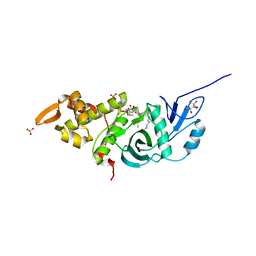 | | Crystal structure of yeast Esa1 HAT domain bound to coenzyme A with active site lysine acetylated | | Descriptor: | CACODYLIC ACID, COENZYME A, GLYCEROL, ... | | Authors: | Yuan, H, Ding, E.C, Marmorstein, R. | | Deposit date: | 2011-09-04 | | Release date: | 2011-11-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | MYST protein acetyltransferase activity requires active site lysine autoacetylation.
Embo J., 31, 2011
|
|
4R5K
 
 | | Crystal structure of the DnaK C-terminus (Dnak-SBD-B) | | Descriptor: | CALCIUM ION, Chaperone protein DnaK, SULFATE ION | | Authors: | Leu, J.I, Zhang, P, Murphy, M.E, Marmorstein, R, George, D.L. | | Deposit date: | 2014-08-21 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7469 Å) | | Cite: | Structural Basis for the Inhibition of HSP70 and DnaK Chaperones by Small-Molecule Targeting of a C-Terminal Allosteric Pocket.
Acs Chem.Biol., 9, 2014
|
|
3TOA
 
 | | Human MOF crystal structure with active site lysine partially acetylated | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, ZINC ION, ... | | Authors: | Yuan, H, Ding, E.C, Marmorstein, R. | | Deposit date: | 2011-09-04 | | Release date: | 2011-11-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.004 Å) | | Cite: | MYST protein acetyltransferase activity requires active site lysine autoacetylation.
Embo J., 31, 2011
|
|
