4KVO
 
 | |
4KVM
 
 | |
7RB3
 
 | |
6UV5
 
 | |
6UUW
 
 | | Structure of human ATP citrate lyase E599Q mutant in complex with Mg2+, citrate, ATP and CoA | | Descriptor: | (2S)-2-hydroxy-2-[2-oxo-2-(phosphonooxy)ethyl]butanedioic acid, ADENOSINE-5'-DIPHOSPHATE, ATP-citrate synthase, ... | | Authors: | Wei, X, Marmorstein, R. | | Deposit date: | 2019-11-01 | | Release date: | 2019-12-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Molecular basis for acetyl-CoA production by ATP-citrate lyase
Nat.Struct.Mol.Biol., 27, 2020
|
|
8GIX
 
 | | Chaetomium thermophilum Hir3 | | Descriptor: | 3,3',3''-phosphanetriyltripropanoic acid, Histone transcription regulator 3 homolog | | Authors: | Szurgot, M.R, van Eeuwen, T, Kim, H.J, Marmorstein, R. | | Deposit date: | 2023-03-14 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the Hir histone chaperone complex.
Mol.Cell, 2024
|
|
1HU8
 
 | | CRYSTAL STRUCTURE OF THE MOUSE P53 CORE DNA-BINDING DOMAIN AT 2.7A RESOLUTION | | Descriptor: | CELLULAR TUMOR ANTIGEN P53, ZINC ION | | Authors: | Zhao, K, Chai, X, Johnston, K, Clements, A, Marmorstein, R. | | Deposit date: | 2001-01-04 | | Release date: | 2001-07-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the mouse p53 core DNA-binding domain at 2.7 A resolution.
J.Biol.Chem., 276, 2001
|
|
1ZME
 
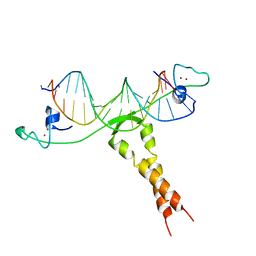 | | CRYSTAL STRUCTURE OF PUT3/DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*CP*GP*GP*AP*GP*(5IU)P*TP*GP*GP*CP*TP*(5IU)P*CP*CP*CP*G)-3'), DNA (5'-D(*AP*CP*GP*GP*GP*AP*AP*GP*CP*CP*AP*AP*CP*TP*CP*CP*G)-3'), PROLINE UTILIZATION TRANSCRIPTION ACTIVATOR, ... | | Authors: | Swaminathan, K, Marmorstein, R. | | Deposit date: | 1997-08-06 | | Release date: | 1998-09-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a PUT3-DNA complex reveals a novel mechanism for DNA recognition by a protein containing a Zn2Cys6 binuclear cluster.
Nat.Struct.Biol., 4, 1997
|
|
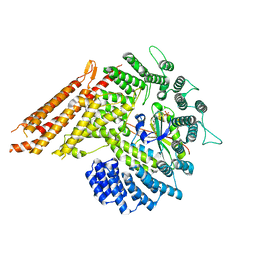
8G0L
 
 | |
4E26
 
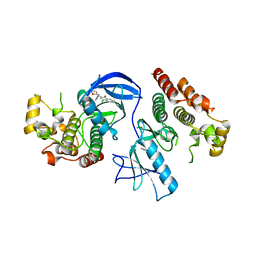 | | BRAF in complex with an organic inhibitor 7898734 | | Descriptor: | 5-chloro-7-[(R)-furan-2-yl(pyridin-2-ylamino)methyl]quinolin-8-ol, Serine/threonine-protein kinase B-raf | | Authors: | Qin, J, Xie, P, Ventocilla, C, Zhou, G, Vultur, A, Chen, Q, Herlyn, M, Winkler, J, Marmorstein, R. | | Deposit date: | 2012-03-07 | | Release date: | 2012-05-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Identification of a Novel Family of BRAF(V600E) Inhibitors.
J.Med.Chem., 55, 2012
|
|
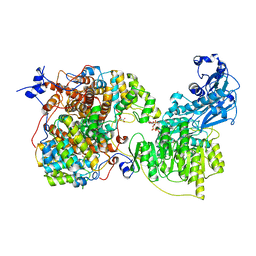
7LJ9
 
 | |
4KVX
 
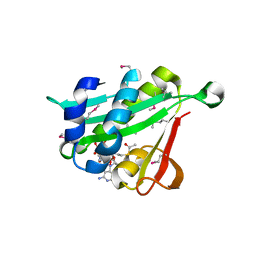 | | Crystal structure of Naa10 (Ard1) bound to AcCoA | | Descriptor: | ACETYL COENZYME *A, N-terminal acetyltransferase A complex catalytic subunit ard1 | | Authors: | Liszczak, G.P, Marmorstein, R.Q. | | Deposit date: | 2013-05-23 | | Release date: | 2013-07-31 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for N-terminal acetylation by the heterodimeric NatA complex.
Nat.Struct.Mol.Biol., 20, 2013
|
|
6C95
 
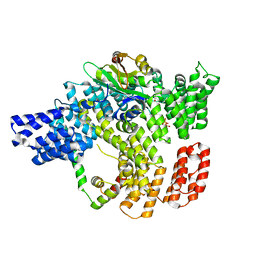 | |
2QIY
 
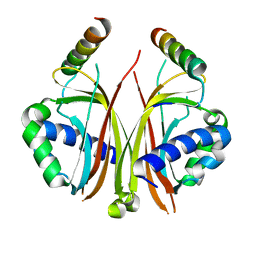 | |
2B9D
 
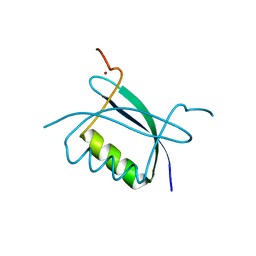 | | Crystal Structure of HPV E7 CR3 domain | | Descriptor: | E7 protein, ZINC ION | | Authors: | Liu, X, Clements, A, Zhao, K, Marmorstein, R. | | Deposit date: | 2005-10-11 | | Release date: | 2005-10-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the human Papillomavirus E7 oncoprotein and its mechanism for inactivation of the retinoblastoma tumor suppressor.
J.Biol.Chem., 281, 2006
|
|
6VP9
 
 | | Cryo-EM structure of human NatB complex | | Descriptor: | CARBOXYMETHYL COENZYME *A, MDVFM peptide, N-alpha-acetyltransferase 20, ... | | Authors: | Deng, S, Marmorstein, R. | | Deposit date: | 2020-02-02 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Molecular basis for N-terminal alpha-synuclein acetylation by human NatB.
Elife, 9, 2020
|
|
3FY0
 
 | |
6C9M
 
 | |
3FXZ
 
 | |
6POF
 
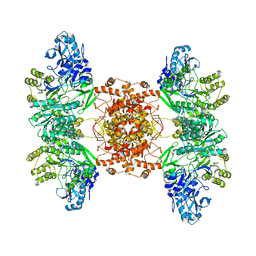 | | Structure of human ATP citrate lyase | | Descriptor: | ATP-citrate synthase | | Authors: | Wei, X, Marmorstein, R. | | Deposit date: | 2019-07-03 | | Release date: | 2020-01-01 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Molecular basis for acetyl-CoA production by ATP-citrate lyase.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6PPL
 
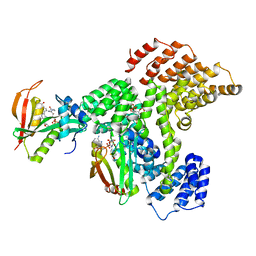 | | Cryo-EM structure of human NatE complex (NatA/Naa50) | | Descriptor: | ACETYL COENZYME *A, INOSITOL HEXAKISPHOSPHATE, N-alpha-acetyltransferase 10, ... | | Authors: | Deng, S, Marmorstein, R. | | Deposit date: | 2019-07-08 | | Release date: | 2020-02-19 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Molecular basis for N-terminal acetylation by human NatE and its modulation by HYPK.
Nat Commun, 11, 2020
|
|
6PW9
 
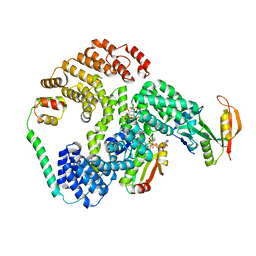 | | Cryo-EM structure of human NatE/HYPK complex | | Descriptor: | ACETYL COENZYME *A, Huntingtin-interacting protein K, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Deng, S, Marmorstein, R. | | Deposit date: | 2019-07-22 | | Release date: | 2020-02-19 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (4.03 Å) | | Cite: | Molecular basis for N-terminal acetylation by human NatE and its modulation by HYPK.
Nat Commun, 11, 2020
|
|
2FQ3
 
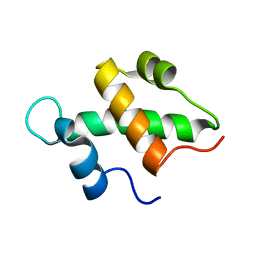 | | Structure and function of the SWIRM domain, a conserved protein module found in chromatin regulatory complexes | | Descriptor: | Transcription regulatory protein SWI3 | | Authors: | Da, G, Lenkart, J, Zhao, K, Shiekhattar, R, Cairns, B.R, Marmorstein, R. | | Deposit date: | 2006-01-17 | | Release date: | 2006-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and function of the SWIRM domain, a conserved protein module found in chromatin regulatory complexes
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
5ICW
 
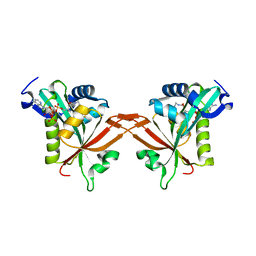 | | Crystal structure of human NatF (hNaa60) homodimer bound to Coenzyme A | | Descriptor: | CHLORIDE ION, COENZYME A, N-alpha-acetyltransferase 60 | | Authors: | Stove, S.I, Magin, R.S, Marmorstein, R, Arnesen, T. | | Deposit date: | 2016-02-23 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Crystal Structure of the Golgi-Associated Human N alpha-Acetyltransferase 60 Reveals the Molecular Determinants for Substrate-Specific Acetylation.
Structure, 24, 2016
|
|
1S5P
 
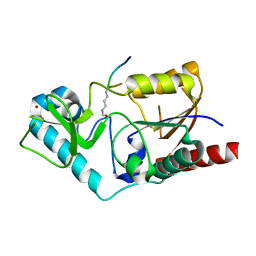 | | Structure and substrate binding properties of cobB, a Sir2 homolog protein deacetylase from Eschericia coli. | | Descriptor: | HISTONE H4 (RESIDUES 12-19), NAD-dependent deacetylase, ZINC ION | | Authors: | Zhao, K, Chai, X, Marmorstein, R. | | Deposit date: | 2004-01-21 | | Release date: | 2004-03-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure and Substrate Binding Properties of cobB, a Sir2 Homolog Protein Deacetylase from Eschericia coli.
J.Mol.Biol., 337, 2004
|
|
