6SV1
 
 | |
6SUW
 
 | |
5N5F
 
 | |
7OE2
 
 | | Model of closed pentamer of the Haliangium ochraceum encapsulin from symmetry expansion of icosahedral single particle reconstruction | | Descriptor: | Haliangium ochraceum Encapsulated ferritin localisation sequence, Linocin_M18 bacteriocin protein | | Authors: | Marles-Wright, J, Basle, A, Clarke, D.J, Ross, J. | | Deposit date: | 2021-05-01 | | Release date: | 2022-02-09 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Pore dynamics and asymmetric cargo loading in an encapsulin nanocompartment.
Sci Adv, 8, 2022
|
|
7ODW
 
 | |
5N5E
 
 | |
7OEU
 
 | | Model of open pentamer of the Haliangium ochraceum encapsulin from symmetry expansion of icosahedral single particle reconstruction | | Descriptor: | Haliangium ochraceum encapsulated ferritin, Linocin_M18 bacteriocin protein | | Authors: | Marles-Wright, J, Basle, A, Clarke, D.J, Ross, J. | | Deposit date: | 2021-05-04 | | Release date: | 2022-02-09 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Pore dynamics and asymmetric cargo loading in an encapsulin nanocompartment.
Sci Adv, 8, 2022
|
|
2XXQ
 
 | | A widespread family of bacterial cell wall assembly proteins | | Descriptor: | (2Z,6Z,10Z,14Z,18Z,22Z,26Z)-3,7,11,15,19,23,27,31-octamethyldotriaconta-2,6,10,14,18,22,26,30-octaen-1-yl trihydrogen diphosphate, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Marles-Wright, J, Kawai, Y, Emmins, R, Ishikawa, S, Kuwano, M, Heinz, N, Cleverley, R.M, Bui, N.K, Ogasawara, N, Lewis, R.J, Vollmer, W, Daniel, R.A, Errington, J. | | Deposit date: | 2010-11-11 | | Release date: | 2011-10-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | A Widespread Family of Bacterial Cell Wall Assembly Proteins.
Embo J., 30, 2011
|
|
2XXP
 
 | | A widespread family of bacterial cell wall assembly proteins | | Descriptor: | CPS2A, DI(HYDROXYETHYL)ETHER, MONO-TRANS, ... | | Authors: | Marles-Wright, J, Kawai, Y, Emmins, R, Ishikawa, S, Kuwano, M, Heinz, N, Cleverley, R.M, Bui, N.K, Ogasawara, N, Lewis, R.J, Vollmer, W, Daniel, R.A, Errington, J. | | Deposit date: | 2010-11-11 | | Release date: | 2011-10-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.692 Å) | | Cite: | A Widespread Family of Bacterial Cell Wall Assembly Proteins.
Embo J., 30, 2011
|
|
2Y0O
 
 | |
5CML
 
 | | Crystal structure of the Esterase domain from Rhodothermus marinus Rmar_1206 protein | | Descriptor: | OsmC family protein, TRIETHYLENE GLYCOL | | Authors: | Marles-Wright, J, Wardrope, C, Jensen, M.-B.V, Horsfall, L.E, Togneri, P.D, Rosser, S.J. | | Deposit date: | 2015-07-16 | | Release date: | 2016-07-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Characterisation of a New Family of Carboxyl Esterases with an OsmC Domain.
Plos One, 11, 2016
|
|
2VY9
 
 | | Molecular architecture of the stressosome, a signal integration and transduction hub | | Descriptor: | ANTI-SIGMA-FACTOR ANTAGONIST | | Authors: | Marles-Wright, J, Grant, T, Delumeau, O, van Duinen, G, Firbank, S.J, Lewis, P.J, Murray, J.W, Newman, J.A, Quin, M.B, Race, P.R, Rohou, A, Tichelaar, W, van Heel, M, Lewis, R.J. | | Deposit date: | 2008-07-21 | | Release date: | 2008-10-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular Architecture of the "Stressosome," a Signal Integration and Transduction Hub
Science, 322, 2008
|
|
4C3S
 
 | | Structure of a propionaldehyde dehydrogenase from the Clostridium phytofermentans fucose utilisation bacterial microcompartment | | Descriptor: | ALDEHYDE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Marles-Wright, J, Crawshaw, A, Ang, T.F, Altenbach, K. | | Deposit date: | 2013-08-27 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Insight Into Coenzyme a Cofactor Binding and the Mechanism of Acyl-Transfer in an Acylating Aldehyde Dehydrogenase from Clostridium Phytofermentans.
Sci.Rep., 6, 2016
|
|
4TQX
 
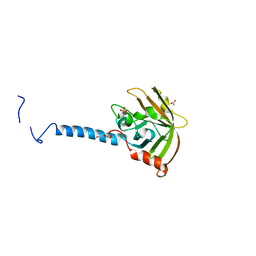 | | Molecular Basis of Streptococcus mutans Sortase A Inhibition by Chalcone. | | Descriptor: | ACETIC ACID, SULFATE ION, Sortase, ... | | Authors: | Wallock-Richards, D.J, Marles-Wright, J, Clarke, D.J, Maitra, A, Dodds, M, Hanley, B, Campopiano, D.J. | | Deposit date: | 2014-06-12 | | Release date: | 2015-05-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Molecular basis of Streptococcus mutans sortase A inhibition by the flavonoid natural product trans-chalcone.
Chem.Commun.(Camb.), 51, 2015
|
|
3TEP
 
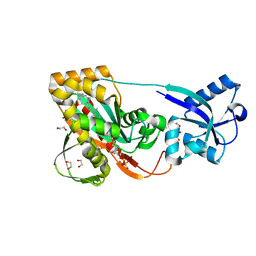 | | LytR-CPS2a-Psr family protein with bound octaprenyl pyrophosphate lipid and magnesium ion | | Descriptor: | (2Z,6Z,10Z,14Z,18Z,22Z,26Z)-3,7,11,15,19,23,27,31-octamethyldotriaconta-2,6,10,14,18,22,26,30-octaen-1-yl trihydrogen diphosphate, 1,2-ETHANEDIOL, CPS2A, ... | | Authors: | Kawai, Y, Marles-Wright, J, Cleverley, R.M, Emmins, R, Ishikawa, S, Kuwano, M, Heinz, N, Khai Bui, N, Hoyland, C.N, Ogasawara, N, Lewis, R.J, Vollmer, W, Daniel, R.A, Errington, J. | | Deposit date: | 2011-08-15 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A widespread family of bacterial cell wall assembly proteins.
Embo J., 30, 2011
|
|
3TEL
 
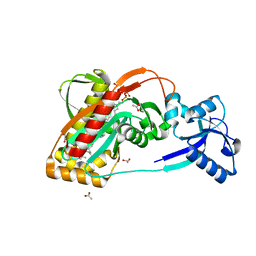 | | LytR-CPS2A-Psr family protein with bound octaprenyl pyrophosphate lipid and manganese ion | | Descriptor: | (2Z,6Z,10Z,14Z,18Z,22Z,26Z)-3,7,11,15,19,23,27,31-octamethyldotriaconta-2,6,10,14,18,22,26,30-octaen-1-yl trihydrogen diphosphate, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Kawai, Y, Marles-Wright, J, Cleverley, R.M, Emmins, R, Ishikawa, S, Kuwano, M, Heinz, N, Khai Bui, N, Hoyland, C.N, Ogasawara, N, Lewis, R.J, Vollmer, W, Daniel, R.A, Errington, J. | | Deposit date: | 2011-08-15 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A widespread family of bacterial cell wall assembly proteins.
Embo J., 30, 2011
|
|
3TFL
 
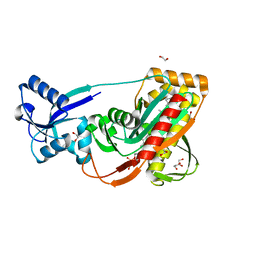 | | LytR-Cps2a-Psr family protein with bound octaprenyl pyrophosphate lipid | | Descriptor: | (2Z,6Z,10Z,14Z,18Z,22Z,26Z)-3,7,11,15,19,23,27,31-octamethyldotriaconta-2,6,10,14,18,22,26,30-octaen-1-yl trihydrogen diphosphate, 1,2-ETHANEDIOL, CPS2A, ... | | Authors: | Kawai, Y, Marles-Wright, J, Cleverley, R.M, Emmins, R, Ishikawa, S, Kuwano, M, Heinz, N, Khai Bui, N, Hoyland, C.N, Ogasawara, N, Lewis, R.J, Vollmer, W, Daniel, R.A, Errington, J. | | Deposit date: | 2011-08-16 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A widespread family of bacterial cell wall assembly proteins.
Embo J., 30, 2011
|
|
7POA
 
 | | An Irreversible, Promiscuous and Highly Thermostable Claisen-Condensation Biocatalyst Drives the Synthesis of Substituted Pyrroles | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 8-amino-7-oxononanoate synthase/2-amino-3-ketobutyrate coenzyme A ligase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Basle, A, Ashley, B, Campopiano, D, Marles-Wright, J. | | Deposit date: | 2021-09-08 | | Release date: | 2022-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Versatile Chemo-Biocatalytic Cascade Driven by a Thermophilic and Irreversible C-C Bond-Forming alpha-Oxoamine Synthase.
Acs Sustain Chem Eng, 11, 2023
|
|
7POB
 
 | | An Irreversible, Promiscuous and Highly Thermostable Claisen-Condensation Biocatalyst Drives the Synthesis of Substituted Pyrroles | | Descriptor: | 8-amino-7-oxononanoate synthase/2-amino-3-ketobutyrate coenzyme A ligase, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION | | Authors: | Basle, A, Ashley, B, Campopiano, D, Marles-Wright, J. | | Deposit date: | 2021-09-08 | | Release date: | 2022-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Versatile Chemo-Biocatalytic Cascade Driven by a Thermophilic and Irreversible C-C Bond-Forming alpha-Oxoamine Synthase.
Acs Sustain Chem Eng, 11, 2023
|
|
7POC
 
 | | An Irreversible, Promiscuous and Highly Thermostable Claisen-Condensation Biocatalyst Drives the Synthesis of Substituted Pyrroles | | Descriptor: | 8-amino-7-oxononanoate synthase/2-amino-3-ketobutyrate coenzyme A ligase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Basle, A, Ashley, B, Campopiano, D, Marles-Wright, J. | | Deposit date: | 2021-09-08 | | Release date: | 2022-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Versatile Chemo-Biocatalytic Cascade Driven by a Thermophilic and Irreversible C-C Bond-Forming alpha-Oxoamine Synthase.
Acs Sustain Chem Eng, 11, 2023
|
|
8C46
 
 | |
6G1F
 
 | |
3QEE
 
 | | The structure and function of an arabinan-specific alpha-1,2-arabinofuranosidase identified from screening the activities of bacterial GH43 glycoside hydrolases | | Descriptor: | ACETATE ION, Beta-xylosidase/alpha-L-arabinfuranosidase, gly43N, ... | | Authors: | Cartmell, A, Mckee, L.S, Pena, M, Larsbrink, J, Brumer, H, Lewis, R.J, Viks-Nielsen, A, Gilbert, H.J, Marles-Wright, J. | | Deposit date: | 2011-01-20 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | The Structure and Function of an Arabinan-specific {alpha}-1,2-Arabinofuranosidase Identified from Screening the Activities of Bacterial GH43 Glycoside Hydrolases.
J.Biol.Chem., 286, 2011
|
|
3QED
 
 | | The structure and function of an arabinan-specific alpha-1,2-arabinofuranosidase identified from screening the activities of bacterial GH43 glycoside hydrolases | | Descriptor: | Beta-xylosidase/alpha-L-arabinfuranosidase, gly43N, CALCIUM ION, ... | | Authors: | Cartmell, A, Mckee, L.S, Pena, M, Larsbrink, J, Brumer, H, Lewis, R.J, Viks-Nielsen, A, Gilbert, H.J, Marles-Wright, J. | | Deposit date: | 2011-01-20 | | Release date: | 2011-02-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The Structure and Function of an Arabinan-specific {alpha}-1,2-Arabinofuranosidase Identified from Screening the Activities of Bacterial GH43 Glycoside Hydrolases.
J.Biol.Chem., 286, 2011
|
|
3QEF
 
 | | The structure and function of an arabinan-specific alpha-1,2-arabinofuranosidase identified from screening the activities of bacterial GH43 glycoside hydrolases | | Descriptor: | 1,2-ETHANEDIOL, Beta-xylosidase/alpha-L-arabinfuranosidase, gly43N, ... | | Authors: | Cartmell, A, Mckee, L.S, Pena, M, Larsbrink, J, Brumer, H, Lewis, R.J, Viks-Nielsen, A, Gilbert, H.J, Marles-Wright, J. | | Deposit date: | 2011-01-20 | | Release date: | 2011-02-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.789 Å) | | Cite: | The Structure and Function of an Arabinan-specific {alpha}-1,2-Arabinofuranosidase Identified from Screening the Activities of Bacterial GH43 Glycoside Hydrolases.
J.Biol.Chem., 286, 2011
|
|
