6UE4
 
 | |
5WUF
 
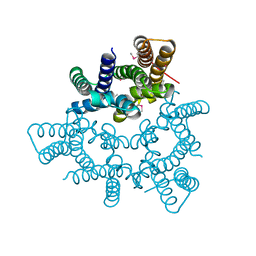 | | Structural basis for conductance through TRIC cation channels | | Descriptor: | CADMIUM ION, Putative membrane protein | | Authors: | Mao, Y, Gao, F, Su, M, Wang, X.H, Zeng, Y, Bruni, R, Kloss, B, Hendrickson, W.A, Chen, Y.H, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2016-12-17 | | Release date: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structural basis for conductance through TRIC cation channels.
Nat Commun, 8, 2017
|
|
6PLM
 
 | | Legionella pneumophila SidJ/ Calmodulin 2 complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, CALCIUM ION, Calmodulin-2, ... | | Authors: | Mao, Y, Sulpizio, A, Minelli, M.E, Wu, X. | | Deposit date: | 2019-07-01 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.592 Å) | | Cite: | Protein polyglutamylation catalyzed by the bacterial calmodulin-dependent pseudokinase SidJ.
Elife, 8, 2019
|
|
6B7O
 
 | | Crystal structure of Legionella effector sdeD (lpg2509) H67A in complex with ADP-ribosylated Ubiquitin | | Descriptor: | Polyubiquitin-C, SdeD (lpg2509) H67A, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Mao, Y, Akturk, A, Wasilko, J. | | Deposit date: | 2017-10-04 | | Release date: | 2018-04-18 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism of phosphoribosyl-ubiquitination mediated by a single Legionella effector.
Nature, 557, 2018
|
|
6B7P
 
 | |
7XGT
 
 | |
2KIG
 
 | |
6B7Q
 
 | |
6B7M
 
 | |
1HX8
 
 | | CRYSTAL STRUCTURE OF N-TERMINAL DOMAIN OF DROSOPHILA AP180 | | Descriptor: | SULFATE ION, SYNAPSE-ENRICHED CLATHRIN ADAPTOR PROTEIN LAP | | Authors: | Mao, Y, Chen, J, Maynard, J.A, Zhang, B, Quiocho, F.A. | | Deposit date: | 2001-01-12 | | Release date: | 2001-02-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A novel all helix fold of the AP180 amino-terminal domain for phosphoinositide binding and clathrin assembly in synaptic vesicle endocytosis.
Cell(Cambridge,Mass.), 104, 2001
|
|
3OGN
 
 | |
1TT9
 
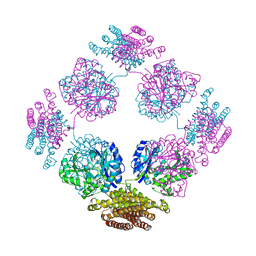 | | Structure of the bifunctional and Golgi associated formiminotransferase cyclodeaminase octamer | | Descriptor: | Formimidoyltransferase-cyclodeaminase (Formiminotransferase- cyclodeaminase) (FTCD) (58 kDa microtubule-binding protein) | | Authors: | Mao, Y, Vyas, N.K, Vyas, M.N, Chen, D.H, Ludtke, S.J, Chiu, W, Quiocho, F.A. | | Deposit date: | 2004-06-22 | | Release date: | 2005-06-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Structure of the bifunctional and Golgi-associated formiminotransferase cyclodeaminase octamer
Embo J., 23, 2004
|
|
1DVP
 
 | | CRYSTAL STRUCTURE OF THE VHS AND FYVE TANDEM DOMAINS OF HRS, A PROTEIN INVOLVED IN MEMBRANE TRAFFICKING AND SIGNAL TRANSDUCTION | | Descriptor: | CITRIC ACID, HEPATOCYTE GROWTH FACTOR-REGULATED TYROSINE KINASE SUBSTRATE, ZINC ION | | Authors: | Mao, Y, Nickitenko, A, Duan, X, Lloyd, T.E, Wu, M.N, Bellen, H, Quiocho, F.A. | | Deposit date: | 2000-01-21 | | Release date: | 2000-03-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the VHS and FYVE tandem domains of Hrs, a protein involved in membrane trafficking and signal transduction.
Cell(Cambridge,Mass.), 100, 2000
|
|
2QV2
 
 | |
3LWT
 
 | | Crystal structure of the Yeast Sac1: Implications for its phosphoinositide phosphatase function | | Descriptor: | Phosphoinositide phosphatase SAC1 | | Authors: | Mao, Y, Manford, A, Xia, T, Saxena, A.K, Stefan, C, Hu, F, Emr, S.D. | | Deposit date: | 2010-02-24 | | Release date: | 2010-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.956 Å) | | Cite: | Crystal structure of the yeast Sac1: implications for its phosphoinositide phosphatase function.
Embo J., 29, 2010
|
|
2AGA
 
 | | De-ubiquitinating function of ataxin-3: insights from the solution structure of the Josephin domain | | Descriptor: | Machado-Joseph disease protein 1 | | Authors: | Mao, Y, Senic-Matuglia, F, Di Fiore, P, Polo, S, Hodsdon, M.E, De Camilli, P. | | Deposit date: | 2005-07-26 | | Release date: | 2005-08-30 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Deubiquitinating function of ataxin-3: insights from the solution structure of the Josephin domain.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2KIE
 
 | | A PH domain within OCRL bridges clathrin mediated membrane trafficking to phosphoinositide metabolis | | Descriptor: | Inositol polyphosphate 5-phosphatase OCRL-1 | | Authors: | Mao, Y, Balkin, D.M, Zoncu, R, Erdmann, K, Tomasini, L, Hu, F, Jin, M.M, Hodsdon, M.E, De Camilli, P. | | Deposit date: | 2009-05-03 | | Release date: | 2009-07-21 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | A PH domain within OCRL bridges clathrin-mediated membrane trafficking to phosphoinositide metabolism
Embo J., 28, 2009
|
|
5WUC
 
 | | Structural basis for conductance through TRIC cation channels | | Descriptor: | SODIUM ION, Uncharacterized protein | | Authors: | Su, M, Gao, F, Mao, Y, Li, D.L, Guo, Y.Z, Wang, X.H, Bruni, R, Kloss, B, Hendrickson, W.A, Chen, Y.H, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2016-12-17 | | Release date: | 2017-07-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for conductance through TRIC cation channels.
Nat Commun, 8, 2017
|
|
5WUD
 
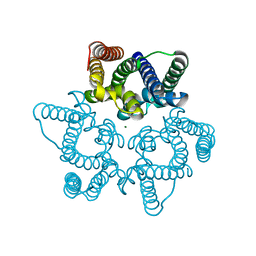 | | Structural basis for conductance through TRIC cation channels | | Descriptor: | MAGNESIUM ION, Uncharacterized protein | | Authors: | Su, M, Gao, F, Mao, Y, Li, D.L, Guo, Y.Z, Wang, X.H, Bruni, R, Kloss, B, Hendrickson, W.A, Chen, Y.H, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2016-12-17 | | Release date: | 2017-06-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for conductance through TRIC cation channels.
Nat Commun, 8, 2017
|
|
5WUE
 
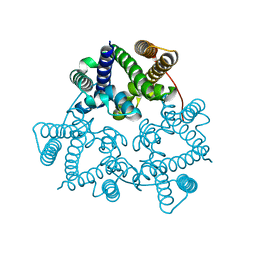 | | Structural basis for conductance through TRIC cation channels | | Descriptor: | SULFATE ION, Uncharacterized protein | | Authors: | Su, M, Gao, F, Mao, Y, Li, D.L, Guo, Y.Z, Wang, X.H, Bruni, R, Kloss, B, Hendrickson, W.A, Chen, Y.H, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2016-12-17 | | Release date: | 2017-06-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for conductance through TRIC cation channels.
Nat Commun, 8, 2017
|
|
5T0I
 
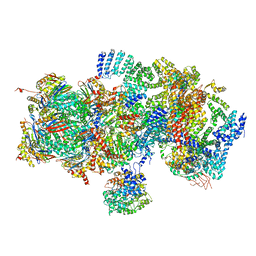 | | Structural basis for dynamic regulation of the human 26S proteasome | | Descriptor: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | Authors: | Chen, S, Wu, J, Lu, Y, Ma, Y.B, Lee, B.H, Yu, Z, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | Deposit date: | 2016-08-16 | | Release date: | 2016-10-19 | | Last modified: | 2016-11-30 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structural basis for dynamic regulation of the human 26S proteasome.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5T0C
 
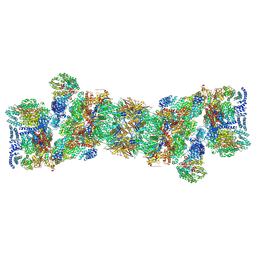 | | Structural basis for dynamic regulation of the human 26S proteasome | | Descriptor: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | Authors: | Chen, S, Wu, J, Lu, Y, Ma, Y.B, Lee, B.H, Yu, Z, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | Deposit date: | 2016-08-15 | | Release date: | 2016-10-19 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for dynamic regulation of the human 26S proteasome.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5T0J
 
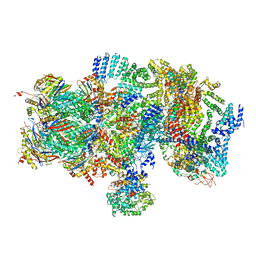 | | Structural basis for dynamic regulation of the human 26S proteasome | | Descriptor: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | Authors: | Chen, S, Wu, J, Lu, Y, Ma, Y.B, Lee, B.H, Yu, Z, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | Deposit date: | 2016-08-16 | | Release date: | 2016-10-19 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structural basis for dynamic regulation of the human 26S proteasome.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5T0G
 
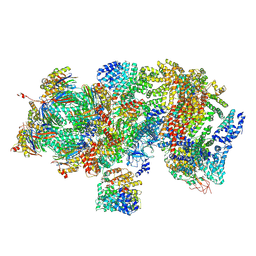 | | Structural basis for dynamic regulation of the human 26S proteasome | | Descriptor: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | Authors: | Chen, S, Wu, J, Lu, Y, Ma, Y.B, Lee, B.H, Yu, Z, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | Deposit date: | 2016-08-16 | | Release date: | 2016-10-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural basis for dynamic regulation of the human 26S proteasome.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5T0H
 
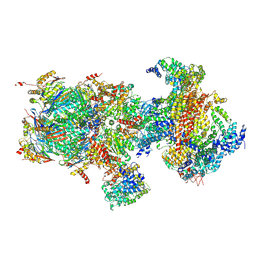 | | Structural basis for dynamic regulation of the human 26S proteasome | | Descriptor: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | Authors: | Chen, S, Wu, J, Lu, Y, Ma, Y.B, Lee, B.H, Yu, Z, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | Deposit date: | 2016-08-16 | | Release date: | 2016-10-19 | | Last modified: | 2016-11-30 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Structural basis for dynamic regulation of the human 26S proteasome.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
