6F2U
 
 | | Potent and selective Aldo-Keto Reductase 1C3 (AKR1C3) inhibitors based on the benzoisoxazole moiety: application of a Bioisosteric Scaffold Hopping Approach to Flufenamic acid | | Descriptor: | 3-[(4-methoxyphenyl)methyl]-5-oxidanyl-~{N}-[3-(trifluoromethyl)phenyl]-1,2,3-triazole-4-carboxamide, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Goyal, P, Wahlgren, W.Y, Friemann, R. | | Deposit date: | 2017-11-27 | | Release date: | 2018-04-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Potent and selective aldo-keto reductase 1C3 (AKR1C3) inhibitors based on the benzoisoxazole moiety: application of a bioisosteric scaffold hopping approach to flufenamic acid.
Eur J Med Chem, 150, 2018
|
|
5MVD
 
 | | Crystal structure of potent human Dihydroorotate Dehydrogenase inhibitors based on hydroxylated azole scaffolds | | Descriptor: | 1,5-dimethyl-3-oxidanyl-~{N}-[2,3,5,6-tetrakis(fluoranyl)-4-phenyl-phenyl]pyrazole-4-carboxamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Goyal, P, Andersson, M, Moritzer, A.C, Sainas, S, Pippione, A.C, Boschi, D, Al-Kadaraghi, S, Lolli, M, Friemann, R. | | Deposit date: | 2017-01-16 | | Release date: | 2017-03-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design, synthesis, biological evaluation and X-ray structural studies of potent human dihydroorotate dehydrogenase inhibitors based on hydroxylated azole scaffolds.
Eur J Med Chem, 129, 2017
|
|
6F78
 
 | | Potent and selective Aldo-Keto Reductase 1C3 (AKR1C3) inhibitors based on the benzoisoxazole moiety: Application of a Bioisosteric Scaffold Hopping Approach to Flufenamic acid | | Descriptor: | 4-[[3,5-bis(trifluoromethyl)phenyl]amino]-1,2-benzoxazol-3-one, Aldo-keto reductase family 1 member C3, CHLORIDE ION, ... | | Authors: | Goyal, P, Wahlgren, W.Y, Friemann, R. | | Deposit date: | 2017-12-07 | | Release date: | 2018-04-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Potent and selective aldo-keto reductase 1C3 (AKR1C3) inhibitors based on the benzoisoxazole moiety: application of a bioisosteric scaffold hopping approach to flufenamic acid.
Eur J Med Chem, 150, 2018
|
|
6G5I
 
 | | Cryo-EM structure of a late human pre-40S ribosomal subunit - State R | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Ameismeier, M, Cheng, J, Berninghausen, O, Beckmann, R. | | Deposit date: | 2018-03-29 | | Release date: | 2018-06-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Visualizing late states of human 40S ribosomal subunit maturation.
Nature, 558, 2018
|
|
6G53
 
 | | Cryo-EM structure of a late human pre-40S ribosomal subunit - State E | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Ameismeier, M, Cheng, J, Berninghausen, O, Beckmann, R. | | Deposit date: | 2018-03-28 | | Release date: | 2018-06-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Visualizing late states of human 40S ribosomal subunit maturation.
Nature, 558, 2018
|
|
6G4S
 
 | | Cryo-EM structure of a late human pre-40S ribosomal subunit - State B | | Descriptor: | 40S ribosomal protein S11, 40S ribosomal protein S13, 40S ribosomal protein S14, ... | | Authors: | Ameismeier, M, Cheng, J, Berninghausen, O, Beckmann, R. | | Deposit date: | 2018-03-28 | | Release date: | 2018-06-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Visualizing late states of human 40S ribosomal subunit maturation.
Nature, 558, 2018
|
|
5NCK
 
 | |
6G5H
 
 | | Cryo-EM structure of a late human pre-40S ribosomal subunit - Mature | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Ameismeier, M, Cheng, J, Berninghausen, O, Beckmann, R. | | Deposit date: | 2018-03-29 | | Release date: | 2018-06-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Visualizing late states of human 40S ribosomal subunit maturation.
Nature, 558, 2018
|
|
6G4W
 
 | | Cryo-EM structure of a late human pre-40S ribosomal subunit - State A | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S11, 40S ribosomal protein S13, ... | | Authors: | Ameismeier, M, Cheng, J, Berninghausen, O, Beckmann, R. | | Deposit date: | 2018-03-28 | | Release date: | 2018-06-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Visualizing late states of human 40S ribosomal subunit maturation.
Nature, 558, 2018
|
|
6G51
 
 | | Cryo-EM structure of a late human pre-40S ribosomal subunit - State D | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Ameismeier, M, Cheng, J, Berninghausen, O, Beckmann, R. | | Deposit date: | 2018-03-28 | | Release date: | 2018-06-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Visualizing late states of human 40S ribosomal subunit maturation.
Nature, 558, 2018
|
|
5MVC
 
 | | Crystal structure of potent human Dihydroorotate Dehydrogenase inhibitors based on hydroxylated azole scaffolds | | Descriptor: | 4-oxidanyl-~{N}-[2,3,5,6-tetrakis(fluoranyl)-4-phenyl-phenyl]-1,2,5-thiadiazole-3-carboxamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Goyal, P, Andersson, M, Moritzer, A.C, Sainas, S, Pippione, A.C, Boschi, D, Al-Kadaraghi, S, Lolli, M, Friemann, R. | | Deposit date: | 2017-01-16 | | Release date: | 2017-03-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Design, synthesis, biological evaluation and X-ray structural studies of potent human dihydroorotate dehydrogenase inhibitors based on hydroxylated azole scaffolds.
Eur J Med Chem, 129, 2017
|
|
5MUT
 
 | | Crystal structure of potent human Dihydroorotate Dehydrogenase inhibitors based on hydroxylated azole scaffolds | | Descriptor: | 2-methyl-5-oxidanyl-~{N}-[2,3,5,6-tetrakis(fluoranyl)-4-phenyl-phenyl]-1,2,3-triazole-4-carboxamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Goyal, P, Andersson, M, Moritzer, A.C, Sainas, S, Pippione, A.C, Boschi, D, Al-Kadaraghi, S, Lolli, M, Friemann, R. | | Deposit date: | 2017-01-14 | | Release date: | 2017-03-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Design, synthesis, biological evaluation and X-ray structural studies of potent human dihydroorotate dehydrogenase inhibitors based on hydroxylated azole scaffolds.
Eur J Med Chem, 129, 2017
|
|
4PJ3
 
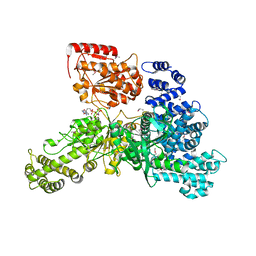 | | Structural insight into the function and evolution of the spliceosomal helicase Aquarius, Structure of Aquarius in complex with AMPPNP | | Descriptor: | Intron-binding protein aquarius, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | De, I, Bessonov, S, Hofele, R, dos Santos, K.F, Will, C.L, Urlaub, H, Luhrmann, R, Pena, V. | | Deposit date: | 2014-05-11 | | Release date: | 2015-01-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The RNA helicase Aquarius exhibits structural adaptations mediating its recruitment to spliceosomes.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5M5H
 
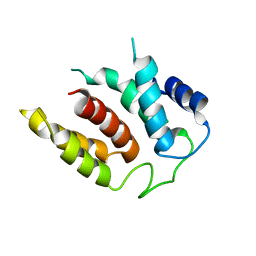 | | RIBOSOME-BOUND YIDC INSERTASE | | Descriptor: | Membrane protein insertase YidC | | Authors: | Kedrov, A, Wickles, S, Crevenna, A.H, van der Sluis, E, Buschauer, R, Berninghausen, O, Lamb, D.C, Beckmann, R. | | Deposit date: | 2016-10-21 | | Release date: | 2016-12-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural Dynamics of the YidC:Ribosome Complex during Membrane Protein Biogenesis.
Cell Rep, 17, 2016
|
|
5NVA
 
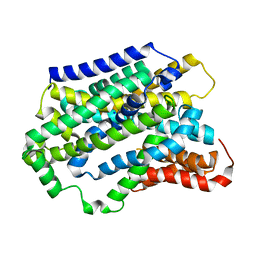 | | Substrate-bound outward-open state of a Na+-coupled sialic acid symporter reveals a novel Na+-site | | Descriptor: | N-acetyl-beta-neuraminic acid, Putative sodium:solute symporter, SODIUM ION | | Authors: | Wahlgren, W.Y, North, R.A, Dunevall, E, Goyal, P, Grabe, M, Dobson, R, Abramson, J, Ramaswamy, S, Friemann, R. | | Deposit date: | 2017-05-03 | | Release date: | 2018-04-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Substrate-bound outward-open structure of a Na+-coupled sialic acid symporter reveals a new Na+site.
Nat Commun, 9, 2018
|
|
6H58
 
 | | Structure of a hibernating 100S ribosome reveals an inactive conformation of the ribosomal protein S1 - Full 100S Hibernating E. coli Ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S1, ... | | Authors: | Beckert, B, Turk, M, Czech, A, Berninghausen, O, Beckmann, R, Ignatova, Z, Plitzko, J, Wilson, D.N. | | Deposit date: | 2018-07-24 | | Release date: | 2018-09-05 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Structure of a hibernating 100S ribosome reveals an inactive conformation of the ribosomal protein S1.
Nat Microbiol, 3, 2018
|
|
6H4N
 
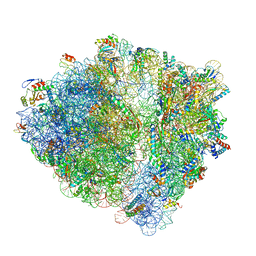 | | Structure of a hibernating 100S ribosome reveals an inactive conformation of the ribosomal protein S1 - 70S Hibernating E. coli Ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S1, ... | | Authors: | Beckert, B, Turk, M, Czech, A, Berninghausen, O, Beckmann, R, Ignatova, Z, Plitzko, J, Wilson, N.D. | | Deposit date: | 2018-07-22 | | Release date: | 2018-09-05 | | Last modified: | 2018-10-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of a hibernating 100S ribosome reveals an inactive conformation of the ribosomal protein S1.
Nat Microbiol, 3, 2018
|
|
6FF4
 
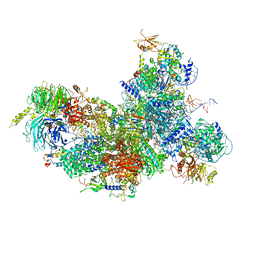 | | human Bact spliceosome core structure | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, BUD13 homolog, Cell division cycle 5-like protein, ... | | Authors: | Haselbach, D, Komarov, I, Agafonov, D, Hartmuth, K, Graf, B, Kastner, B, Luehrmann, R, Stark, H. | | Deposit date: | 2018-01-03 | | Release date: | 2018-08-29 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure and Conformational Dynamics of the Human Spliceosomal BactComplex.
Cell, 172, 2018
|
|
5LQW
 
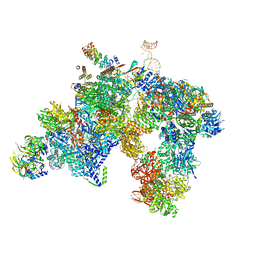 | | yeast activated spliceosome | | Descriptor: | Pre-mRNA leakage protein 1, Pre-mRNA-processing protein 45, Pre-mRNA-splicing factor 8, ... | | Authors: | Rauhut, R, Luehrmann, R. | | Deposit date: | 2016-08-17 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Molecular architecture of the Saccharomyces cerevisiae activated spliceosome
Science, 6306, 2016
|
|
6HD5
 
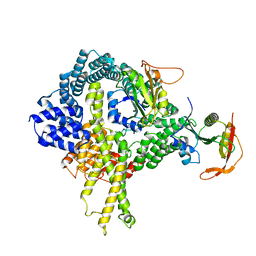 | | Cryo-EM structure of the ribosome-NatA complex | | Descriptor: | N-alpha-acetyltransferase NAT5, N-terminal acetyltransferase A complex catalytic subunit ARD1, N-terminal acetyltransferase A complex subunit NAT1 | | Authors: | Knorr, A.G, Becker, T, Beckmann, R. | | Deposit date: | 2018-08-17 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Ribosome-NatA architecture reveals that rRNA expansion segments coordinate N-terminal acetylation.
Nat. Struct. Mol. Biol., 26, 2019
|
|
6GXK
 
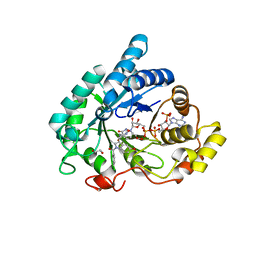 | | Crystal structure of Aldo-Keto Reductase 1C3 (AKR1C3) complexed with inhibitor. | | Descriptor: | 1,2-ETHANEDIOL, 4-[[1-(4-chlorophenyl)carbonyl-5-methoxy-2-methyl-indol-3-yl]methyl]-1,2,5-oxadiazol-3-one, Aldo-keto reductase family 1 member C3, ... | | Authors: | Goyal, P, Wahlgren, W.Y, Friemann, R. | | Deposit date: | 2018-06-27 | | Release date: | 2019-05-08 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Bioisosteres of Indomethacin as Inhibitors of Aldo-Keto Reductase 1C3.
Acs Med.Chem.Lett., 10, 2019
|
|
7OJ0
 
 | | Cryo-EM structure of 70S ribosome stalled with TnaC peptide and RF2 | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Su, T, Kudva, R, Becker, T, Berninghausen, O, Heijne, G, Cheng, J, Beckmann, R. | | Deposit date: | 2021-05-13 | | Release date: | 2021-09-15 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of l-tryptophan-dependent inhibition of release factor 2 by the TnaC arrest peptide.
Nucleic Acids Res., 49, 2021
|
|
7OIZ
 
 | | Cryo-EM structure of 70S ribosome stalled with TnaC peptide | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Su, T, Kudva, R, Becker, T, Berninghausen, O, Heijne, G, Cheng, J, Beckmann, R. | | Deposit date: | 2021-05-13 | | Release date: | 2021-09-15 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of l-tryptophan-dependent inhibition of release factor 2 by the TnaC arrest peptide.
Nucleic Acids Res., 49, 2021
|
|
5LW7
 
 | | S. solfataricus ABCE1 post-splitting state | | Descriptor: | ABC transporter ATP-binding protein, IRON/SULFUR CLUSTER | | Authors: | Heuer, A, Gerovac, M, Beckmann, R, Tampe, R. | | Deposit date: | 2016-09-15 | | Release date: | 2016-11-16 | | Last modified: | 2019-02-20 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Structure of the ribosome post-recycling complex probed by chemical cross-linking and mass spectrometry.
Nat Commun, 7, 2016
|
|
8QPP
 
 | | Bacillus subtilis MutS2-collided disome complex (stalled 70S) | | Descriptor: | 16S rRNA (1533-MER), 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Park, E, Mackens-Kiani, T, Berhane, R, Esser, H, Erdenebat, C, Burroughs, A.M, Berninghausen, O, Aravind, L, Beckmann, R, Green, R, Buskirk, A.R. | | Deposit date: | 2023-10-02 | | Release date: | 2023-12-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | B. subtilis MutS2 splits stalled ribosomes into subunits without mRNA cleavage.
Embo J., 43, 2024
|
|
