3KK5
 
 | |
1MVM
 
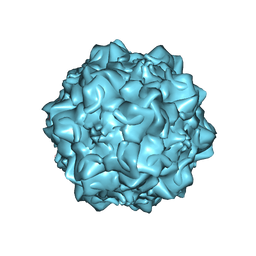 | | MVM(STRAIN I), COMPLEX(VIRAL COAT/DNA), VP2, PH=7.5, T=4 DEGREES C | | Descriptor: | DNA (5'-D(*CP*AP*AP*A)-3'), DNA (5'-D(*CP*CP*AP*CP*CP*CP*CP*AP*AP*CP*A)-3'), DNA (5'-D(P*A)-3'), ... | | Authors: | Llamas-Saiz, A.L, Agbandje-McKenna, M, Rossmann, M.G. | | Deposit date: | 1996-06-21 | | Release date: | 1998-02-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure determination of minute virus of mice.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
1QJX
 
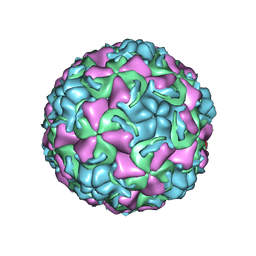 | | HUMAN RHINOVIRUS 16 COAT PROTEIN IN COMPLEX WITH ANTIVIRAL COMPOUND WIN68934 | | Descriptor: | 2,6-DIMETHYL-1-(3-[3-METHYL-5-ISOXAZOLYL]-PROPANYL)-4-[4-METHYL-2H-TETRAZOL-2-YL]-PHENOL, MYRISTIC ACID, PROTEIN VP1, ... | | Authors: | Hadfield, A.T, Diana, G.D, Rossmann, M.G. | | Deposit date: | 1999-07-06 | | Release date: | 1999-07-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Analysis of Three Structurally Related Antiviral Compounds in Complex with Human Rhinovirus 16
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
2A08
 
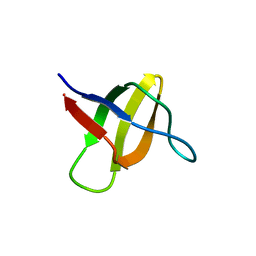 | | Structure of the yeast YHH6 SH3 domain | | Descriptor: | Hypothetical 41.8 kDa protein in SPO13-ARG4 intergenic region | | Authors: | Kursula, P, Kursula, I, Song, Y.H, Lehmann, F, Zou, P, Wilmanns, M. | | Deposit date: | 2005-06-16 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | 3-D proteome of yeast SH3 domains
To be Published
|
|
5CYI
 
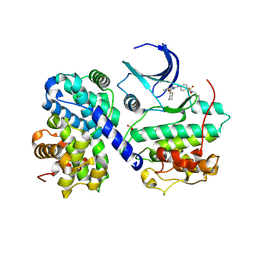 | | CDK2/Cyclin A covalent complex with 6-(cyclohexylmethoxy)-N-(4-(vinylsulfonyl)phenyl)-9H-purin-2-amine (NU6300) | | Descriptor: | 6-(cyclohexylmethoxy)-N-[4-(ethylsulfonyl)phenyl]-9H-purin-2-amine, Cyclin-A2, Cyclin-dependent kinase 2 | | Authors: | Anscombe, E, Meschini, E, Vidal, R.M, Martin, M.P, Staunton, D, Geitmann, M, Danielson, U.H, Stanley, W.A, Wang, L.Z, Reuillon, T, Golding, B.T, Cano, C, Newell, D.R, Noble, M.E.M, Wedge, S.R, Endicott, J.A, Griffin, R.J. | | Deposit date: | 2015-07-30 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification and Characterization of an Irreversible Inhibitor of CDK2.
Chem.Biol., 22, 2015
|
|
1TG8
 
 | | The structure of Dengue virus E glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, envelope glycoprotein | | Authors: | Zhang, Y, Zhang, W, Ogata, S, Clements, D, Strauss, J.H, Baker, T.S, Rossmann, M.G. | | Deposit date: | 2004-05-28 | | Release date: | 2004-09-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Conformational changes of the flavivirus e glycoprotein
Structure, 12, 2004
|
|
1TGE
 
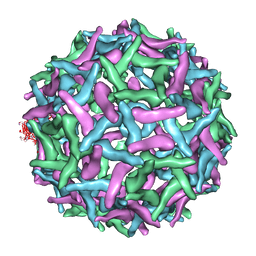 | | The structure of immature Dengue virus at 12.5 angstrom | | Descriptor: | envelope glycoprotein | | Authors: | Zhang, Y, Zhang, W, Ogata, S, Clements, D, Strauss, J.H, Baker, T.S, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2004-05-28 | | Release date: | 2004-09-28 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12.5 Å) | | Cite: | Conformational changes of the flavivirus e glycoprotein.
Structure, 12, 2004
|
|
4USH
 
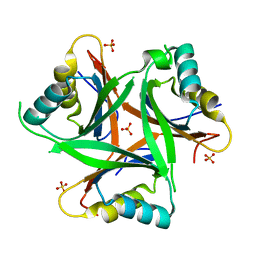 | |
1ND3
 
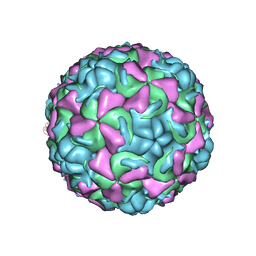 | | The structure of HRV16, when complexed with pleconaril, an antiviral compound | | Descriptor: | 3-{3,5-DIMETHYL-4-[3-(3-METHYL-ISOXAZOL-5-YL)-PROPOXY]-PHENYL}-5-TRIFLUOROMETHYL-[1,2,4]OXADIAZOLE, ZINC ION, coat protein VP1, ... | | Authors: | Zhang, Y, Simpson, A.A, Bator, C.M, Chakravarty, S, Pevear, D.C, Skochko, G.A, Tull, T.M, Diana, G, Rossmann, M.G. | | Deposit date: | 2002-12-06 | | Release date: | 2003-12-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and virological studies of the stages of virus replication that are affected by antirhinovirus compounds
J.Virol., 78, 2004
|
|
2YKL
 
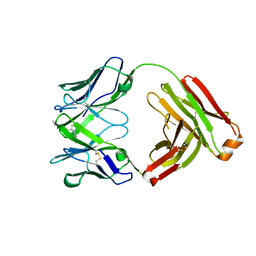 | | Structure of human anti-nicotine Fab fragment in complex with nicotine-11-yl-methyl-(4-ethylamino-4-oxo)-butanoate | | Descriptor: | FAB FRAGMENT, HEAVY CHAIN, LIGHT CHAIN, ... | | Authors: | Tars, K, Kotelovica, S, Lipowsky, G, Bauer, M, Beerli, R, Bachmann, M, Maurer, P. | | Deposit date: | 2011-05-27 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Different Binding Modes of Free and Carrier-Protein-Coupled Nicotine in a Human Monoclonal Antibody.
J.Mol.Biol., 415, 2012
|
|
1ND2
 
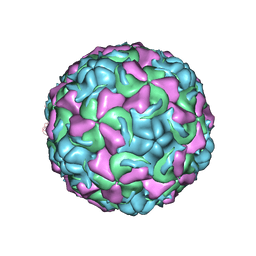 | | The structure of Rhinovirus 16 | | Descriptor: | MYRISTIC ACID, ZINC ION, coat protein VP1, ... | | Authors: | Zhang, Y, Simpson, A.A, Bator, C.M, Chakravarty, S, Pevear, D.C, Skochko, G.A, Tull, T.M, Diana, G, Rossmann, M.G. | | Deposit date: | 2002-12-06 | | Release date: | 2003-12-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and virological studies of the stages of virus replication that are affected by antirhinovirus compounds
J.Virol., 78, 2004
|
|
1NCQ
 
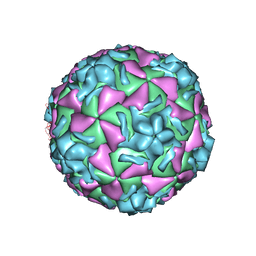 | | The structure of HRV14 when complexed with pleconaril, an antiviral compound | | Descriptor: | 3-{3,5-DIMETHYL-4-[3-(3-METHYL-ISOXAZOL-5-YL)-PROPOXY]-PHENYL}-5-TRIFLUOROMETHYL-[1,2,4]OXADIAZOLE, COAT PROTEIN VP1, COAT PROTEIN VP2, ... | | Authors: | Zhang, Y, Simpson, A.A, Bator, C.M, Chakravarty, S, Pevear, D.C, Skochko, G.A, Tull, T.M, Diana, G, Rossmann, M.G. | | Deposit date: | 2002-12-05 | | Release date: | 2003-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and virological studies of the stages of virus replication that are affected by antirhinovirus compounds
J.Virol., 78, 2004
|
|
2Y9M
 
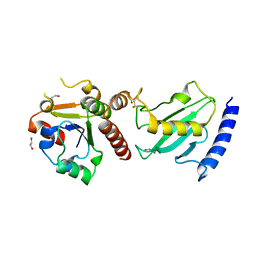 | | Pex4p-Pex22p structure | | Descriptor: | 1,2-ETHANEDIOL, PEROXISOME ASSEMBLY PROTEIN 22, UBIQUITIN-CONJUGATING ENZYME E2-21 KDA | | Authors: | Williams, C, van den Berg, M, Panjikar, S, Distel, B, Wilmanns, M. | | Deposit date: | 2011-02-15 | | Release date: | 2011-10-26 | | Last modified: | 2017-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insights Into Ubiquitin-Conjugating Enzyme/ Co-Activator Interactions from the Structure of the Pex4P:Pex22P Complex.
Embo J., 31, 2011
|
|
7AC8
 
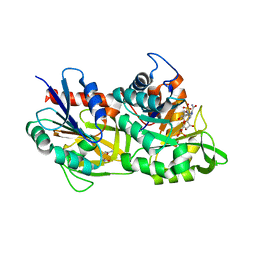 | |
3IY0
 
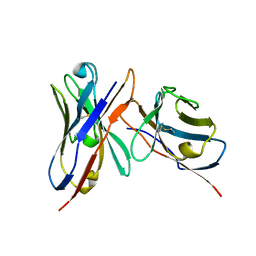 | | Variable domains of the x-ray structure of Fab 14 fitted into the cryoEM reconstruction of the virus-Fab 14 complex | | Descriptor: | Fab 14, heavy domain, light domain | | Authors: | Hafenstein, S, Bowman, V.D, Sun, T, Nelson, C.D, Palermo, L.M, Chipman, P.R, Battisti, A.J, Parrish, C.R, Rossmann, M.G. | | Deposit date: | 2009-04-07 | | Release date: | 2009-05-12 | | Last modified: | 2011-07-13 | | Method: | ELECTRON MICROSCOPY (12.5 Å) | | Cite: | Structural comparison of different antibodies interacting with parvovirus capsids
J.Virol., 83, 2009
|
|
4WM8
 
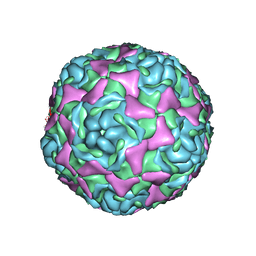 | | Crystal Structure of Human Enterovirus D68 | | Descriptor: | DECANOIC ACID, VP1, VP2, ... | | Authors: | Liu, Y, Sheng, J, Fokine, A, Meng, G, Long, F, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2014-10-08 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Virus structure. Structure and inhibition of EV-D68, a virus that causes respiratory illness in children.
Science, 347, 2015
|
|
1PDI
 
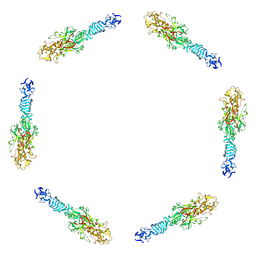 | | Fitting of the C-terminal part of the short tail fibers into the cryo-EM reconstruction of T4 baseplate | | Descriptor: | Short tail fiber protein | | Authors: | Kostyuchenko, V.A, Leiman, P.G, Chipman, P.R, Kanamaru, S, van Raaij, M.J, Arisaka, F, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2003-05-19 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Three-dimensional structure of bacteriophage T4 baseplate
Nat.Struct.Biol., 10, 2003
|
|
1BWT
 
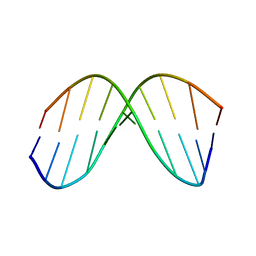 | |
3IY7
 
 | | Variable domains of the computer generated model (WAM) of Fab F fitted into the cryoEM reconstruction of the virus-Fab F complex | | Descriptor: | fragment from neutralizing antibody F (heavy chain), fragment from neutralizing antibody F (light chain) | | Authors: | Hafenstein, S, Bowman, V.D, Sun, T, Nelson, C.D, Palermo, L.M, Chipman, P.R, Battisti, A.J, Parrish, C.R, Rossmann, M.G. | | Deposit date: | 2009-04-09 | | Release date: | 2009-05-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | Structural comparison of different antibodies interacting with parvovirus capsids
J.Virol., 83, 2009
|
|
1NA1
 
 | | The structure of HRV14 when complexed with Pleconaril | | Descriptor: | 3-{3,5-DIMETHYL-4-[3-(3-METHYL-ISOXAZOL-5-YL)-PROPOXY]-PHENYL}-5-TRIFLUOROMETHYL-[1,2,4]OXADIAZOLE, Coat protein VP1, Coat protein VP2, ... | | Authors: | Zhang, Y, Simpson, A.A, Bator, C.M, Chakravarty, S, Pevear, D.C, Skochko, G.A, Tull, T.M, Diana, G, Rossmann, M.G. | | Deposit date: | 2002-11-26 | | Release date: | 2003-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural and virological studies of the stages of virus replication that are affected by antirhinovirus compounds
J.Virol., 78, 2004
|
|
1S2E
 
 | | BACTERIOPHAGE T4 GENE PRODUCT 9 (GP9), THE TRIGGER OF TAIL CONTRACTION AND THE LONG TAIL FIBERS CONNECTOR, ALTERNATIVE FIT OF THE FIRST 19 RESIDUES | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Baseplate structural protein Gp9 | | Authors: | Kostyuchenko, V.A, Navruzbekov, G.A, Kurochkina, L.P, Strelkov, S.V, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2004-01-08 | | Release date: | 2004-01-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Structure of Bacteriophage T4 Gene Product 9: The Trigger for Tail Contraction
Structure Fold.Des., 7, 1999
|
|
2HWB
 
 | | A comparison of the anti-rhinoviral drug binding pocket in hrv14 and hrv1a | | Descriptor: | 5-(3-(2,6-DICHLORO-4-(4,5-DIHYDRO-2-OXAZOLYL)PHENOXY)PROPYL)-3-METHYL ISOXAZOLE, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Kim, K.H, Rossmann, M.G. | | Deposit date: | 1994-01-25 | | Release date: | 1994-11-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A comparison of the anti-rhinoviral drug binding pocket in HRV14 and HRV1A.
J.Mol.Biol., 230, 1993
|
|
2HWF
 
 | |
2HWC
 
 | | A COMPARISON OF THE ANTI-RHINOVIRAL DRUG BINDING POCKET IN HRV14 AND HRV1A | | Descriptor: | 5-(5-(2,6-DICHLORO-4-(4,5-DIHYDRO-2-OXAZOLY)PHENOXY)PENTYL)-3-METHYL ISOXAZOLE, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Kim, K.H, Rossmann, M.G. | | Deposit date: | 1994-01-25 | | Release date: | 1994-11-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A comparison of the anti-rhinoviral drug binding pocket in HRV14 and HRV1A.
J.Mol.Biol., 230, 1993
|
|
2HWD
 
 | | A COMPARISON OF THE ANTI-RHINOVIRAL DRUG BINDING POCKET IN HRV14 AND HRV1A | | Descriptor: | 5-(3-(2,6-DICHLORO-4-(4,5-DIHYDRO-2-OXAZOLYL)PHENOXY)PROPYL)-3-METHYL ISOXAZOLE, HUMAN RHINOVIRUS 1A COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 1A COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Kim, K.H, Rossmann, M.G. | | Deposit date: | 1994-01-25 | | Release date: | 1994-09-30 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | A comparison of the anti-rhinoviral drug binding pocket in HRV14 and HRV1A.
J.Mol.Biol., 230, 1993
|
|
