1C8E
 
 | |
1C8F
 
 | | FELINE PANLEUKOPENIA VIRUS EMPTY CAPSID STRUCTURE | | Descriptor: | CALCIUM ION, FELINE PANLEUKOPENIA VIRUS CAPSID | | Authors: | Rossmann, M.G, Simpson, A.A. | | Deposit date: | 2000-05-05 | | Release date: | 2000-08-09 | | Last modified: | 2023-04-19 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Host range and variability of calcium binding by surface loops in the capsids of canine and feline parvoviruses.
J.Mol.Biol., 300, 2000
|
|
1C8G
 
 | | FELINE PANLEUKOPENIA VIRUS EMPTY CAPSID STRUCTURE | | Descriptor: | CALCIUM ION, FELINE PANLEUKOPENIA VIRUS CAPSID | | Authors: | Rossmann, M.G, Simpson, A.A. | | Deposit date: | 2000-05-05 | | Release date: | 2000-08-09 | | Last modified: | 2023-04-19 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Host range and variability of calcium binding by surface loops in the capsids of canine and feline parvoviruses.
J.Mol.Biol., 300, 2000
|
|
1C8D
 
 | |
1C8H
 
 | |
1CD3
 
 | | PROCAPSID OF BACTERIOPHAGE PHIX174 | | Descriptor: | PROTEIN (CAPSID PROTEIN GPF), PROTEIN (SCAFFOLDING PROTEIN GPB), PROTEIN (SCAFFOLDING PROTEIN GPD), ... | | Authors: | Rossmann, M.G, Dokland, T. | | Deposit date: | 1999-03-05 | | Release date: | 1999-04-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The role of scaffolding proteins in the assembly of the small, single-stranded DNA virus phiX174.
J.Mol.Biol., 288, 1999
|
|
1BTN
 
 | | STRUCTURE OF THE BINDING SITE FOR INOSITOL PHOSPHATES IN A PH DOMAIN | | Descriptor: | BETA-SPECTRIN, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE | | Authors: | Wilmanns, M, Hyvoenen, M, Saraste, M. | | Deposit date: | 1995-08-23 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the binding site for inositol phosphates in a PH domain.
EMBO J., 14, 1995
|
|
2M37
 
 | | Structure of lasso peptide astexin-1 | | Descriptor: | ASTEXIN-1 | | Authors: | Zimmermann, M, Hegemann, J.D, Xie, X, Marahiel, M.A. | | Deposit date: | 2013-01-14 | | Release date: | 2013-05-01 | | Method: | SOLUTION NMR | | Cite: | The astexin-1 lasso peptides: biosynthesis, stability, and structural studies.
Chem.Biol., 20, 2013
|
|
1CYW
 
 | | QUINOL OXIDASE (PERIPLASMIC FRAGMENT OF SUBUNIT II) (CYOA) | | Descriptor: | CYOA | | Authors: | Wilmanns, M, Lappalainen, P, Kelly, M, Sauer-Eriksson, E, Saraste, M. | | Deposit date: | 1995-08-22 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the membrane-exposed domain from a respiratory quinol oxidase complex with an engineered dinuclear copper center.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
2MLJ
 
 | |
1C2Q
 
 | |
3MML
 
 | | Allophanate Hydrolase Complex from Mycobacterium smegmatis, Msmeg0435-Msmeg0436 | | Descriptor: | Allophanate hydrolase subunit 1, Allophanate hydrolase subunit 2, CHLORIDE ION | | Authors: | Kaufmann, M, Chernishof, I, Shin, A, Germano, D, Sawaya, M.R, Waldo, G.S, Arbing, M.A, Perry, J, Eisenberg, D, Integrated Center for Structure and Function Innovation (ISFI), TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-04-20 | | Release date: | 2010-04-28 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Allphanate Hydrolase Complex from M. smegmatis, Msmeg0435-Msmeg0436
To be Published
|
|
3N2J
 
 | | Azurin H117G, oxidized form | | Descriptor: | Azurin, COPPER (II) ION | | Authors: | Hoffmann, M, Alagaratnam, S, Canters, G.W, Einsle, O. | | Deposit date: | 2010-05-18 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Probing the reactivity of different forms of azurin by flavin photoreduction.
Febs J., 278, 2011
|
|
3O21
 
 | | High resolution structure of GluA3 N-terminal domain (NTD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 3, PHOSPHATE ION | | Authors: | Rossmann, M, Sukumaran, M, Penn, A.C, Veprintsev, D.B, Babu, M.M, Jensen, M.H, Greger, I.H. | | Deposit date: | 2010-07-22 | | Release date: | 2011-03-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Dynamics and allosteric potential of the AMPA receptor N-terminal domain
Embo J., 30, 2011
|
|
1WDX
 
 | | Yeast BBC1 SH3 domain, triclinic crystal form | | Descriptor: | Myosin tail region-interacting protein MTI1 | | Authors: | Wilmanns, M, Consani Textor, L, Kursula, P, Kursula, I, Lehmann, F, Song, Y.H. | | Deposit date: | 2004-05-19 | | Release date: | 2005-05-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Yeast BBC1 SH3 domain, triclinic crystal form
To be Published
|
|
3MPW
 
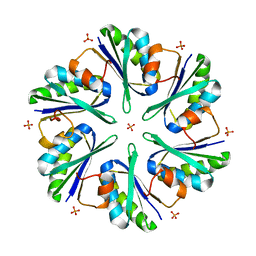 | | Structure of EUTM in 2-D protein membrane | | Descriptor: | Ethanolamine utilization protein eutM, PHOSPHATE ION | | Authors: | Sagermann, M, Takenoya, M, Nikolakakis, K. | | Deposit date: | 2010-04-27 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallographic insights into the pore structures and mechanisms of the EutL and EutM shell proteins of the ethanolamine-utilizing microcompartment of Escherichia coli.
J.Bacteriol., 192, 2010
|
|
3N6V
 
 | |
3MPY
 
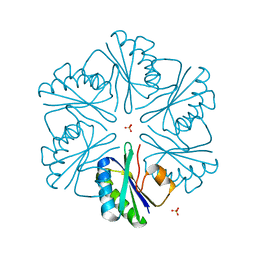 | | Structure of EUTM in 2-D protein membrane | | Descriptor: | Ethanolamine utilization protein eutM, SULFATE ION | | Authors: | Sagermann, M, Takenoya, M, Nikolakakis, K. | | Deposit date: | 2010-04-27 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic insights into the pore structures and mechanisms of the EutL and EutM shell proteins of the ethanolamine-utilizing microcompartment of Escherichia coli.
J.Bacteriol., 192, 2010
|
|
3MPV
 
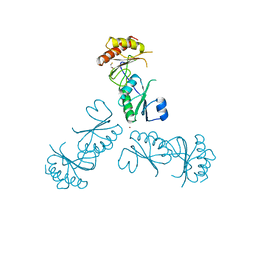 | | Structure of EUTL in the zinc-induced open form | | Descriptor: | BETA-MERCAPTOETHANOL, Ethanolamine utilization protein eutL, ZINC ION | | Authors: | Sagermann, M, Takenoya, M, Nikolakakis, K. | | Deposit date: | 2010-04-27 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystallographic insights into the pore structures and mechanisms of the EutL and EutM shell proteins of the ethanolamine-utilizing microcompartment of Escherichia coli.
J.Bacteriol., 192, 2010
|
|
2MEV
 
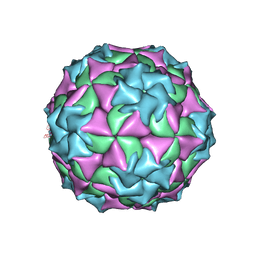 | | STRUCTURAL REFINEMENT AND ANALYSIS OF MENGO VIRUS | | Descriptor: | MENGO VIRUS COAT PROTEIN (SUBUNIT VP1), MENGO VIRUS COAT PROTEIN (SUBUNIT VP2), MENGO VIRUS COAT PROTEIN (SUBUNIT VP3), ... | | Authors: | Rossmann, M.G. | | Deposit date: | 1989-04-21 | | Release date: | 1990-01-15 | | Last modified: | 2023-04-19 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural refinement and analysis of Mengo virus.
J.Mol.Biol., 211, 1990
|
|
2WG6
 
 | | Proteasome-Activating Nucleotidase (PAN) N-domain (57-134) from Archaeoglobus fulgidus fused to GCN4, P61A Mutant | | Descriptor: | GENERAL CONTROL PROTEIN GCN4, PROTEASOME-ACTIVATING NUCLEOTIDASE | | Authors: | Hartmann, M.D, Djuranovic, S, Ursinus, A, Zeth, K, Lupas, A.N. | | Deposit date: | 2009-04-15 | | Release date: | 2009-04-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Activity of the N-Terminal Substrate Recognition Domains in Proteasomal Atpases.
Mol.Cell, 34, 2009
|
|
3O2J
 
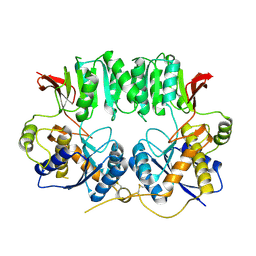 | | Structure of the GluA2 NTD-dimer interface mutant, N54A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 2 | | Authors: | Rossmann, M, Sukumaran, M, Penn, A.C, Veprintsev, D.B, Greger, I.H. | | Deposit date: | 2010-07-22 | | Release date: | 2011-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Subunit-selective N-terminal domain associations organize the formation of AMPA receptor heteromers
Embo J., 30, 2011
|
|
1XRZ
 
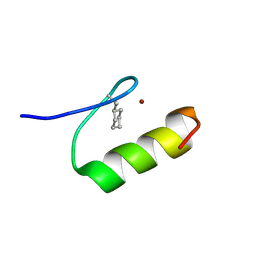 | | NMR Structure of a Zinc Finger with Cyclohexanylalanine Substituted for the Central Aromatic Residue | | Descriptor: | ZINC ION, Zinc finger Y-chromosomal protein | | Authors: | Lachenmann, M.J, Ladbury, J.E, Qian, X, Huang, K, Singh, R, Weiss, M.A. | | Deposit date: | 2004-10-17 | | Release date: | 2004-11-30 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Solvation and the hidden thermodynamics of a zinc finger probed
by nonstandard repair of a protein crevice
Protein Sci., 13, 2004
|
|
3IYW
 
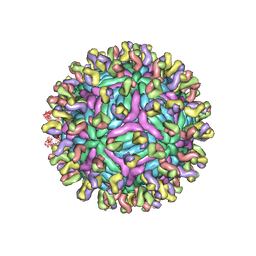 | |
1XF7
 
 | | High Resolution NMR Structure of the Wilms' Tumor Suppressor Protein (WT1) Finger 3 | | Descriptor: | Wilms' Tumor Protein, ZINC ION | | Authors: | Lachenmann, M.J, Ladbury, J.E, Dong, J, Huang, K, Carey, P, Weiss, M.A. | | Deposit date: | 2004-09-14 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Why zinc fingers prefer zinc: ligand-field symmetry and the hidden thermodynamics of metal ion selectivity
Biochemistry, 43, 2004
|
|
