6Z5U
 
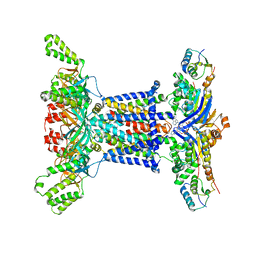 | |
7YYO
 
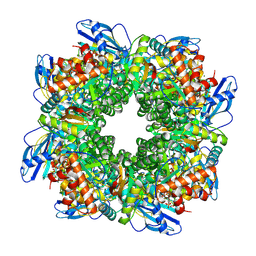 | | Cryo-EM structure of an a-carboxysome RuBisCO enzyme at 2.9 A resolution | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase large chain, ... | | Authors: | Mann, D, Evans, S.L, Bergeron, J.R.C. | | Deposit date: | 2022-02-18 | | Release date: | 2023-01-25 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Single-particle cryo-EM analysis of the shell architecture and internal organization of an intact alpha-carboxysome.
Structure, 31, 2023
|
|
8AFQ
 
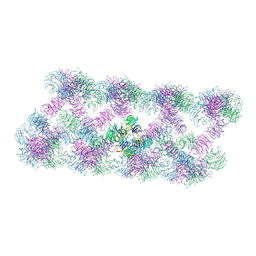 | | Tube assembly of Atg18-PR72AA | | Descriptor: | Autophagy-related protein 18 | | Authors: | Mann, D, Fromm, S, Martinez-Sanchez, A, Gopaldass, N, Mayer, A, Sachse, C. | | Deposit date: | 2022-07-18 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Atg18 oligomer organization in assembled tubes and on lipid membrane scaffolds.
Nat Commun, 14, 2023
|
|
8AFW
 
 | | Tube assembly of Atg18-WT | | Descriptor: | Autophagy-related protein 18 | | Authors: | Mann, D, Fromm, S, Martinez-Sanchez, A, Gopaldass, N, Mayer, A, Sachse, C. | | Deposit date: | 2022-07-18 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Atg18 oligomer organization in assembled tubes and on lipid membrane scaffolds.
Nat Commun, 14, 2023
|
|
8AFX
 
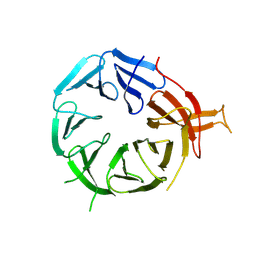 | | Single particle structure of Atg18-WT | | Descriptor: | Autophagy-related protein 18 | | Authors: | Mann, D, Fromm, S, Martinez-Sanchez, A, Gopaldass, N, Mayer, A, Sachse, C. | | Deposit date: | 2022-07-18 | | Release date: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Cryo-EM structures of Atg18 oligomers reveal a tilted structural scaffold for Atg2 at the isolation membrane
To Be Published
|
|
8AFY
 
 | | Subtomogram average of membrane-bound Atg18 oligomers | | Descriptor: | Autophagy-related protein 18 | | Authors: | Mann, D, Fromm, S, Martinez-Sanchez, A, Gopaldass, N, Mayer, A, Sachse, C. | | Deposit date: | 2022-07-18 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (26 Å) | | Cite: | Atg18 oligomer organization in assembled tubes and on lipid membrane scaffolds.
Nat Commun, 14, 2023
|
|
4GS4
 
 | | Structure of the alpha-tubulin acetyltransferase, alpha-TAT1 | | Descriptor: | ACETYL COENZYME *A, Alpha-tubulin N-acetyltransferase | | Authors: | Friedmann, D.R, Fan, J, Marmorstein, R. | | Deposit date: | 2012-08-27 | | Release date: | 2012-10-17 | | Last modified: | 2013-08-28 | | Method: | X-RAY DIFFRACTION (2.112 Å) | | Cite: | Structure of the alpha-tubulin acetyltransferase, alpha TAT1, and implications for tubulin-specific acetylation.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3MKD
 
 | | Crystal structure of myosin-2 dictyostelium discoideum motor domain S456Y mutant in complex with adp-orthovanadate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Myosin-2 heavy chain, ... | | Authors: | Kathmann, D, Diensthuber, R.P, Fedorov, R, Manstein, D.J, Tsiavaliaris, G. | | Deposit date: | 2010-04-14 | | Release date: | 2011-04-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Switch-2 dependent modulation of the myosin power stroke
To be Published
|
|
7M1W
 
 | |
4P7H
 
 | |
4PA0
 
 | | Omecamtiv Mercarbil binding site on the Human Beta-Cardiac Myosin Motor Domain | | Descriptor: | GLYCEROL, Myosin-7,Green fluorescent protein, methyl 4-(2-fluoro-3-{[(6-methylpyridin-3-yl)carbamoyl]amino}benzyl)piperazine-1-carboxylate | | Authors: | Winkelmann, D.A, Miller, M.T, Stock, A.M. | | Deposit date: | 2014-04-06 | | Release date: | 2015-07-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for drug-induced allosteric changes to human beta-cardiac myosin motor activity.
Nat Commun, 6, 2015
|
|
1XXM
 
 | | The modular architecture of protein-protein binding site | | Descriptor: | Beta-lactamase TEM, Beta-lactamase inhibitory protein, CALCIUM ION | | Authors: | Reichmann, D, Rahat, O, Albeck, S, Meged, R, Dym, O, Schreiber, G, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2004-11-07 | | Release date: | 2005-01-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The modular architecture of protein-protein binding interfaces
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1VSG
 
 | |
3IAG
 
 | | CSL (RBP-Jk) bound to HES-1 nonconsensus site | | Descriptor: | 1,2-ETHANEDIOL, 5'-D(*AP*AP*TP*CP*TP*TP*TP*CP*AP*CP*AP*CP*GP*AP*T)-3', 5'-D(*TP*TP*AP*TP*CP*GP*TP*GP*TP*GP*AP*AP*AP*GP*A)-3', ... | | Authors: | Friedmann, D.R, Kovall, R.A. | | Deposit date: | 2009-07-13 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Thermodynamic and structural insights into CSL-DNA complexes.
Protein Sci., 19, 2010
|
|
3NG1
 
 | | N AND GTPASE DOMAINS OF THE SIGNAL SEQUENCE RECOGNITION PROTEIN FFH FROM THERMUS AQUATICUS | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, SIGNAL SEQUENCE RECOGNITION PROTEIN FFH, ... | | Authors: | Freymann, D.M, Stroud, R.M, Walter, P. | | Deposit date: | 1998-09-13 | | Release date: | 1999-07-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Functional changes in the structure of the SRP GTPase on binding GDP and Mg2+GDP.
Nat.Struct.Biol., 6, 1999
|
|
6Y98
 
 | | Crystal Structure of subtype-switched Epithelial Adhesin 9 to 1 A domain (Epa9-CBL2Epa1) from Candida glabrata in complex with beta-lactose | | Descriptor: | CALCIUM ION, PA14 domain-containing protein, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Hoffmann, D, Diderrich, R, Kock, M, Friederichs, S, Reithofer, V, Essen, L.-O, Moesch, H.-U. | | Deposit date: | 2020-03-06 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Functional reprogramming ofCandida glabrataepithelial adhesins: the role of conserved and variable structural motifs in ligand binding.
J.Biol.Chem., 295, 2020
|
|
6Y9J
 
 | | Crystal Structure of subtype-switched Epithelial Adhesin 1 to 9 A domain (Epa1-CBL2Epa9) from Candida glabrata in complex with beta-lactose | | Descriptor: | CALCIUM ION, CHLORIDE ION, Epa1p, ... | | Authors: | Hoffmann, D, Diderrich, R, Kock, M, Friederichs, S, Reithofer, V, Essen, L.-O, Moesch, H.-U. | | Deposit date: | 2020-03-09 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Functional reprogramming ofCandida glabrataepithelial adhesins: the role of conserved and variable structural motifs in ligand binding.
J.Biol.Chem., 295, 2020
|
|
3BRG
 
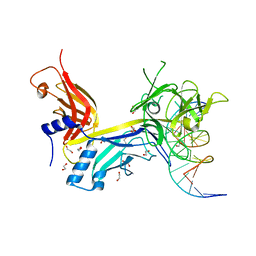 | | CSL (RBP-Jk) bound to DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*DAP*DAP*DTP*DCP*DTP*DTP*DTP*DCP*DCP*DCP*DAP*DCP*DAP*DGP*DT)-3'), DNA (5'-D(*DTP*DTP*DAP*DCP*DTP*DGP*DTP*DGP*DGP*DGP*DAP*DAP*DAP*DGP*DA)-3'), ... | | Authors: | Friedmann, D.R, Kovall, R.A. | | Deposit date: | 2007-12-21 | | Release date: | 2008-04-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | RAM-induced Allostery Facilitates Assembly of a Notch Pathway Active Transcription Complex.
J.Biol.Chem., 283, 2008
|
|
2J46
 
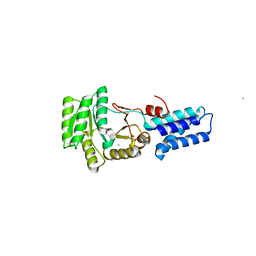 | | Water structure of T. Aquaticus Ffh NG Domain At 1.1A Resolution | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Freymann, D.M, Ramirez, U.D. | | Deposit date: | 2006-08-24 | | Release date: | 2006-11-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Analysis of Protein Hydration in Ultra-High Resolution Structures of the Srp Gtpase Ffh
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2J45
 
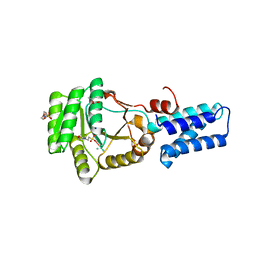 | | Water structure of T. Aquaticus Ffh NG Domain At 1.1A Resolution | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Freymann, D.M, Ramirez, U.D. | | Deposit date: | 2006-08-24 | | Release date: | 2006-11-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Analysis of Protein Hydration in Ultra-High Resolution Structures of the Srp Gtpase Ffh
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2NG1
 
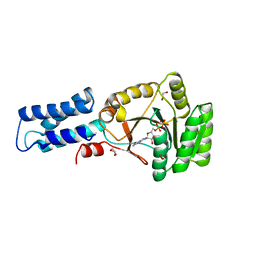 | | N AND GTPASE DOMAINS OF THE SIGNAL SEQUENCE RECOGNITION PROTEIN FFH FROM THERMUS AQUATICUS | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Freymann, D.M, Stroud, R.M, Walter, P. | | Deposit date: | 1998-09-11 | | Release date: | 1999-07-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Functional changes in the structure of the SRP GTPase on binding GDP and Mg2+GDP.
Nat.Struct.Biol., 6, 1999
|
|
2J7P
 
 | |
7QU5
 
 | | X-ray structure of FAD domain of NqrF of Pseudomonas aeruginosa | | Descriptor: | DIMETHYL SULFOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Stegmann, D, Steuber, J, Fritz, G. | | Deposit date: | 2022-01-17 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Fast fragment- and compound-screening pipeline at the Swiss Light Source.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QTY
 
 | | X-ray structure of FAD domain of NqrF of Klebsiella pneumoniae | | Descriptor: | 1-(furan-2-ylmethyl)-3-(2-methylphenyl)thiourea, DIMETHYL SULFOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Stegmann, D, Steuber, J, Fritz, G. | | Deposit date: | 2022-01-17 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Fast fragment- and compound-screening pipeline at the Swiss Light Source.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QU3
 
 | | X-ray structure of FAD domain of NqrF of Pseudomonas aeruginosa | | Descriptor: | 4-(benzimidazol-1-ylmethyl)benzenecarbonitrile, DIMETHYL SULFOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Stegmann, D, Steuber, J, Fritz, G. | | Deposit date: | 2022-01-17 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fast fragment- and compound-screening pipeline at the Swiss Light Source.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
