7K3G
 
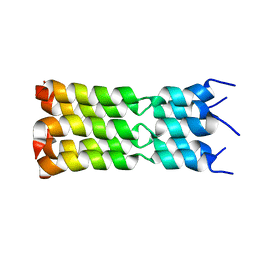 | | SARS-CoV-2 Envelope Protein Transmembrane Domain: Pentameric Structure Determined by Solid-State NMR | | Descriptor: | Envelope small membrane protein | | Authors: | Mandala, V.S, Hong, M, McKay, M.J, Shcherbakov, A.S, Dregni, A.J. | | Deposit date: | 2020-09-11 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Structure and drug binding of the SARS-CoV-2 envelope protein transmembrane domain in lipid bilayers.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6PVR
 
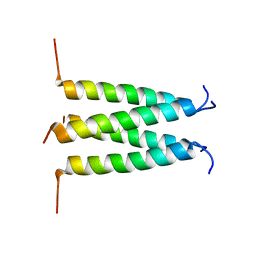 | | Influenza B M2 Proton Channel in the Closed State - SSNMR Structure at pH 7.5 | | Descriptor: | BM2 protein | | Authors: | Mandala, V.S, Loftis, A.R, Shcherbakov, A.S, Pentelute, B.L, Hong, M. | | Deposit date: | 2019-07-21 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Atomic structures of closed and open influenza B M2 proton channel reveal the conduction mechanism.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6PVT
 
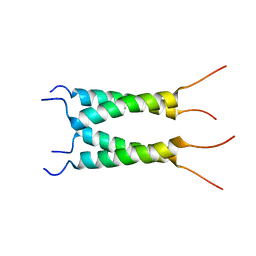 | | Influenza B M2 Proton Channel in the Open State - SSNMR Structure at pH 4.5 | | Descriptor: | BM2 protein | | Authors: | Mandala, V.S, Loftis, A.R, Shcherbakov, A.S, Pentelute, B.L, Hong, M. | | Deposit date: | 2019-07-21 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Atomic structures of closed and open influenza B M2 proton channel reveal the conduction mechanism.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8EP1
 
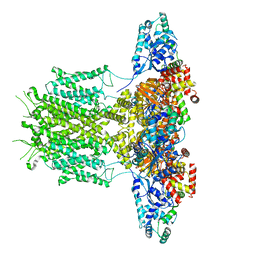 | |
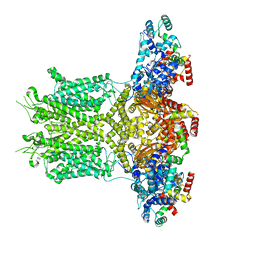
8EOW
 
 | |
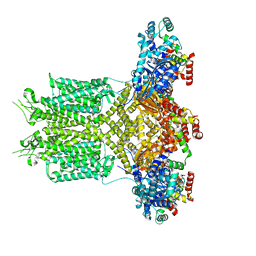
8EP0
 
 | |
8SIK
 
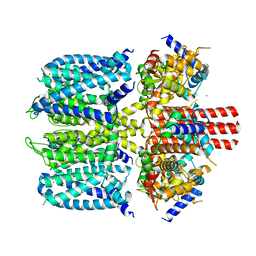 | | KCNQ1 with voltage sensor in the up conformation | | Descriptor: | CALCIUM ION, Calmodulin-1, Potassium voltage-gated channel subfamily KQT member 1 | | Authors: | Mandala, V.S, MacKinnon, R. | | Deposit date: | 2023-04-16 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The membrane electric field regulates the PIP 2 -binding site to gate the KCNQ1 channel.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8SIM
 
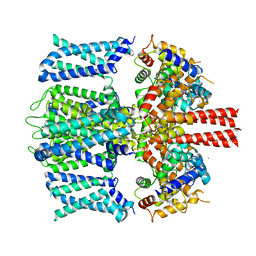 | | KCNQ1 with voltage sensor in the intermediate conformation | | Descriptor: | CALCIUM ION, Calmodulin-1, Potassium voltage-gated channel subfamily KQT member 1 | | Authors: | Mandala, V.S, MacKinnon, R. | | Deposit date: | 2023-04-16 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | The membrane electric field regulates the PIP 2 -binding site to gate the KCNQ1 channel.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8SIN
 
 | | KCNQ1 with voltage sensor in the down conformation | | Descriptor: | Calmodulin-1, Potassium voltage-gated channel subfamily KQT member 1 | | Authors: | Mandala, V.S, MacKinnon, R. | | Deposit date: | 2023-04-16 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | The membrane electric field regulates the PIP 2 -binding site to gate the KCNQ1 channel.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6NZN
 
 | | Dimer-of-dimer amyloid fibril structure of glucagon | | Descriptor: | Glucagon | | Authors: | Gelenter, M.D, Smith, K.J, Liao, S.Y, Mandala, V.S, Dregni, A.J, Lamm, M.S, Tian, Y, Wei, X, Pochan, D.J, Tucker, T.J, Su, Y, Hong, M. | | Deposit date: | 2019-02-14 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | The peptide hormone glucagon forms amyloid fibrils with two coexisting beta-strand conformations.
Nat.Struct.Mol.Biol., 26, 2019
|
|
7JK8
 
 | | EmrE S64V mutant bound to tetra(4-fluorophenyl)phosphonium at pH 5.8 | | Descriptor: | Multidrug SMR transporter, tetrakis(4-fluorophenyl)phosphanium | | Authors: | Shcherbakov, A.A, Hisao, G, Mandala, V.S, Thomas, N.E, Soltani, M, Salter, E.A, Davis Jr, J.H, Henzler-Wildman, K.A, Hong, M. | | Deposit date: | 2020-07-27 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Structure and dynamics of the drug-bound bacterial transporter EmrE in lipid bilayers.
Nat Commun, 12, 2021
|
|
5ZXE
 
 | | Structure of a consensus sequence derived from the FGF family | | Descriptor: | CHLORIDE ION, Consensus sequence based basic form of fibroblast growth factor, GLYCEROL, ... | | Authors: | Tripathi, S.K, Mandalaparthy, V, Ramaswamy, S, Gosavi, S. | | Deposit date: | 2018-05-19 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of a consensus sequence derived from the FGF family
To be published
|
|
