1AVL
 
 | |
1ATN
 
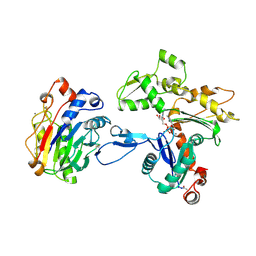 | | Atomic structure of the actin:DNASE I complex | | Descriptor: | ACTIN, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Kabsch, W, Mannherz, H.G, Suck, D, Pai, E, Holmes, K.C. | | Deposit date: | 1991-03-08 | | Release date: | 1992-07-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Atomic structure of the actin:DNase I complex.
Nature, 347, 1990
|
|
7NMS
 
 | |
6N6I
 
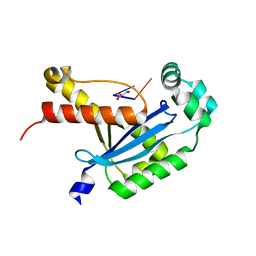 | | Human REXO2 bound to pGG | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, MALONATE ION, ... | | Authors: | Lormand, J.D, Sondermann, H. | | Deposit date: | 2018-11-26 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.431 Å) | | Cite: | A dedicated diribonucleotidase resolves a key bottleneck for the terminal step of RNA degradation.
Elife, 8, 2019
|
|
1BFO
 
 | |
6SLT
 
 | |
4JVF
 
 | | The Crystal structure of PDE6D in complex with the inhibitor (s)-5 | | Descriptor: | (2S)-2-(2-phenyl-1H-benzimidazol-1-yl)-2-(piperidin-4-yl)ethyl 1-(1-benzyl-1H-benzimidazol-2-yl)piperidine-4-carboxylate, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta | | Authors: | Gunther, Z, Papke, B, Ismail, S, Vartak, N, Chandra, A, Hoffmann, M, Hahn, S, Triola, G, Wittinghofer, A, Bastiaens, P, Waldmann, H. | | Deposit date: | 2013-03-25 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Small molecule inhibition of the KRAS PDEd interaction impairs oncogenic KRAS signalling
Nature, 497, 2013
|
|
1RUN
 
 | | CATABOLITE GENE ACTIVATOR PROTEIN (CAP)/DNA COMPLEX + ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, DNA (5'-D(*CP*TP*AP*GP*AP*TP*CP*AP*CP*AP*TP*TP*TP*TP*TP*CP*G )-3'), DNA (5'-D(*GP*CP*GP*AP*AP*AP*AP*AP*TP*GP*TP*GP*AP*T)-3'), ... | | Authors: | Parkinson, G.N, Gunasekera, A, Vojtechovsky, J, Zhang, X, Kunkel, T.A, Berman, H.M, Ebright, R.H. | | Deposit date: | 1996-05-26 | | Release date: | 1997-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Aromatic hydrogen bond in sequence-specific protein DNA recognition.
Nat.Struct.Biol., 3, 1996
|
|
1RUO
 
 | | CATABOLITE GENE ACTIVATOR PROTEIN (CAP) MUTANT/DNA COMPLEX + ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, DNA (5'-D(*CP*TP*AP*GP*AP*TP*CP*AP*CP*AP*TP*TP*TP*TP*TP*CP*G )-3'), DNA (5'-D(*GP*CP*GP*AP*AP*AP*AP*AP*TP*GP*TP*GP*AP*T)-3'), ... | | Authors: | Parkinson, G.N, Ebright, R.H, Berman, H.M. | | Deposit date: | 1996-05-26 | | Release date: | 1997-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Aromatic hydrogen bond in sequence-specific protein DNA recognition.
Nat.Struct.Biol., 3, 1996
|
|
2CDN
 
 | | Crystal structure of Mycobacterium tuberculosis adenylate kinase complexed with two molecules of ADP and Mg | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENYLATE KINASE, MAGNESIUM ION | | Authors: | Bellinzoni, M, Haouz, A, Grana, M, Munier-Lehmann, H, Alzari, P.M. | | Deposit date: | 2006-01-25 | | Release date: | 2006-05-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal Structure of Mycobacterium Tuberculosis Adenylate Kinase in Complex with Two Molecules of Adp and Mg2+ Supports an Associative Mechanism for Phosphoryl Transfer.
Protein Sci., 15, 2006
|
|
6RBD
 
 | | State 1 of yeast Tsr1-TAP Rps20-Deltaloop pre-40S particles | | Descriptor: | 20S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Shayan, R, Mitterer, V, Ferreira-Cerca, S, Murat, G, Enne, T, Rinaldi, D, Weigl, S, Omanic, H, Gleizes, P.E, Kressler, D, Pertschy, B, Plisson-Chastang, C. | | Deposit date: | 2019-04-10 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Conformational proofreading of distant 40S ribosomal subunit maturation events by a long-range communication mechanism.
Nat Commun, 10, 2019
|
|
6RBE
 
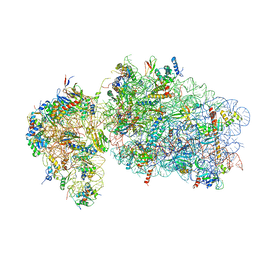 | | State 2 of yeast Tsr1-TAP Rps20-Deltaloop pre-40S particles | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Shayan, R, Mitterer, V, Ferreira-Cerca, S, Murat, G, Enne, T, Rinaldi, D, Weigl, S, Omanic, H, Gleizes, P.E, Kressler, D, Pertschy, B, Plisson-Chastang, C. | | Deposit date: | 2019-04-10 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Conformational proofreading of distant 40S ribosomal subunit maturation events by a long-range communication mechanism.
Nat Commun, 10, 2019
|
|
6RXS
 
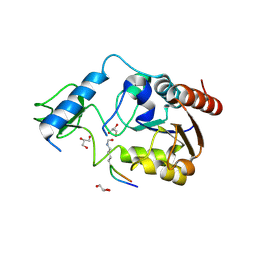 | | Crystal structure of CobB Ac3(A76G,Y92A, I131L, V187Y) in complex with H4K16-Acetyl peptide | | Descriptor: | GLYCEROL, Histone H4, NAD-dependent protein deacylase, ... | | Authors: | Spinck, M, Gasper, R, Neumann, H. | | Deposit date: | 2019-06-08 | | Release date: | 2020-04-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Evolved, Selective Erasers of Distinct Lysine Acylations.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
7QIJ
 
 | |
7QIH
 
 | |
4JV6
 
 | | The crystal structure of PDE6D in complex to inhibitor-1 | | Descriptor: | 1-benzyl-2-phenyl-1H-benzimidazole, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta | | Authors: | Gunther, Z, Papke, B, Ismail, S, Vartak, N, Chandra, A, Hoffmann, M, Hahn, S, Triola, G, Wittinghofer, A, Bastiaens, P, Waldmann, H. | | Deposit date: | 2013-03-25 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Small molecule inhibition of the KRAS PDEd interaction impairs oncogenic KRAS signalling
Nature, 497, 2013
|
|
1MJH
 
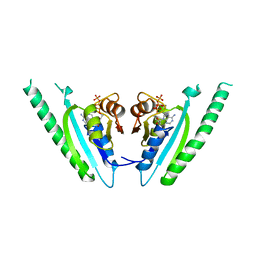 | | Structure-based assignment of the biochemical function of hypothetical protein MJ0577: A test case of structural genomics | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, PROTEIN (ATP-BINDING DOMAIN OF PROTEIN MJ0577) | | Authors: | Zarembinski, T.I, Hung, L.-W, Mueller-Dieckmann, H.J, Kim, K.-K, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 1998-11-04 | | Release date: | 1998-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based assignment of the biochemical function of a hypothetical protein: a test case of structural genomics.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
6RFU
 
 | | In cellulo crystallization of Trypanosoma brucei IMP dehydrogenase enables the identification of ATP and GMP as genuine co-factors | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-MONOPHOSPHATE, Inosine-5'-monophosphate dehydrogenase | | Authors: | Nass, K, Redecke, L, Perbandt, M, Yefanov, O, Gabdulkhakov, A, Duszenko, M, Chapman, H.N, Betzel, C. | | Deposit date: | 2019-04-16 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | In cellulo crystallization of Trypanosoma brucei IMP dehydrogenase enables the identification of genuine co-factors.
Nat Commun, 11, 2020
|
|
6RXM
 
 | | Crystal structure of CobB Ac2 (A76G, I131C, V162G) in complex with H4K16-Acetyl peptide | | Descriptor: | Histone H4, NAD-dependent protein deacylase, ZINC ION | | Authors: | Spinck, M, Gasper, R, Neumann, H. | | Deposit date: | 2019-06-08 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Evolved, Selective Erasers of Distinct Lysine Acylations.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
4JVB
 
 | | Crystal structure of PDE6D in complex with the inhibitor rac-2 | | Descriptor: | 1-benzyl-2-(4-{[(2R)-2-(2-phenyl-1H-benzimidazol-1-yl)pent-4-en-1-yl]oxy}phenyl)-1H-benzimidazole, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta | | Authors: | Gunther, Z, Papke, B, Ismail, S, Vartak, N, Chandra, A, Hoffmann, M, Hahn, S, Triola, G, Wittinghofer, A, Bastiaens, P, Waldmann, H. | | Deposit date: | 2013-03-25 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Small molecule inhibition of the KRAS PDEd interaction impairs oncogenic KRAS signalling
Nature, 497, 2013
|
|
7OL3
 
 | | Human ATL1 N417ins (catalytic core) | | Descriptor: | Atlastin-1, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kelly, C.M, Sondermann, H. | | Deposit date: | 2021-05-19 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A novel insertion mutation in atlastin 1 is associated with spastic quadriplegia, increased membrane tethering, and aberrant conformational switching.
J.Biol.Chem., 298, 2021
|
|
1UK6
 
 | | Crystal structure of a meta-cleavage product hydrolase (CumD) complexed with propionate | | Descriptor: | 2-hydroxy-6-oxo-7-methylocta-2,4-dienoate hydrolase, PROPANOIC ACID | | Authors: | Fushinobu, S, Jun, S.-Y, Hidaka, M, Nojiri, H, Yamane, H, Shoun, H, Omori, T, Wakagi, T. | | Deposit date: | 2003-08-19 | | Release date: | 2004-09-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Series of Crystal Structures of a meta-Cleavage Product Hydrolase from Pseudomonas fluorescens IP01 (CumD) Complexed with Various Cleavage Products
BIOSCI.BIOTECHNOL.BIOCHEM., 69, 2005
|
|
1UK8
 
 | | Crystal structure of a meta-cleavage product hydrolase (CumD) complexed with n-valerate | | Descriptor: | 2-hydroxy-6-oxo-7-methylocta-2,4-dienoate hydrolase, PENTANOIC ACID | | Authors: | Fushinobu, S, Jun, S.-Y, Hidaka, M, Nojiri, H, Yamane, H, Shoun, H, Omori, T, Wakagi, T. | | Deposit date: | 2003-08-19 | | Release date: | 2004-09-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Series of Crystal Structures of a meta-Cleavage Product Hydrolase from Pseudomonas fluorescens IP01 (CumD) Complexed with Various Cleavage Products
BIOSCI.BIOTECHNOL.BIOCHEM., 69, 2005
|
|
1UK7
 
 | | Crystal structure of a meta-cleavage product hydrolase (CumD) complexed with n-butyrate | | Descriptor: | 2-hydroxy-6-oxo-7-methylocta-2,4-dienoate hydrolase, butanoic acid | | Authors: | Fushinobu, S, Jun, S.-Y, Hidaka, M, Nojiri, H, Yamane, H, Shoun, H, Omori, T, Wakagi, T. | | Deposit date: | 2003-08-19 | | Release date: | 2004-09-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Series of Crystal Structures of a meta-Cleavage Product Hydrolase from Pseudomonas fluorescens IP01 (CumD) Complexed with Various Cleavage Products
BIOSCI.BIOTECHNOL.BIOCHEM., 69, 2005
|
|
1UKA
 
 | | Crystal structure of a meta-cleavage product hydrolase (CumD) complexed with (S)-2-methylbutyrate | | Descriptor: | 2-METHYLBUTANOIC ACID, 2-hydroxy-6-oxo-7-methylocta-2,4-dienoate hydrolase | | Authors: | Fushinobu, S, Jun, S.-Y, Hidaka, M, Nojiri, H, Yamane, H, Shoun, H, Omori, T, Wakagi, T. | | Deposit date: | 2003-08-19 | | Release date: | 2004-09-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Series of Crystal Structures of a meta-Cleavage Product Hydrolase from Pseudomonas fluorescens IP01 (CumD) Complexed with Various Cleavage Products
BIOSCI.BIOTECHNOL.BIOCHEM., 69, 2005
|
|
