2JG4
 
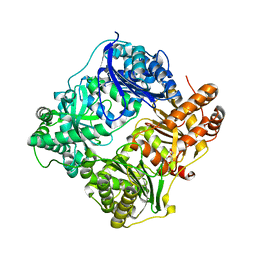 | | Substrate-free IDE structure in its closed conformation | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, INSULIN DEGRADING ENZYME, ZINC ION | | Authors: | Malito, E, Tang, W.J. | | Deposit date: | 2007-02-07 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Substrate-Free Human Insulin Degrading Enzyme (Ide) and Biophysical Analysis of ATP-Induced Conformational Switch of Ide
J.Biol.Chem., 282, 2007
|
|
2YPV
 
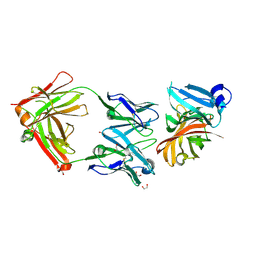 | | Crystal structure of the Meningococcal vaccine antigen factor H binding protein in complex with a bactericidal antibody | | Descriptor: | 1,2-ETHANEDIOL, FAB 12C1, LIPOPROTEIN | | Authors: | Malito, E, Veggi, D, Bottomley, M.J. | | Deposit date: | 2012-11-01 | | Release date: | 2013-02-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Defining a Protective Epitope on Factor H Binding Protein, a Key Meningococcal Virulence Factor and Vaccine Antigen.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1W4X
 
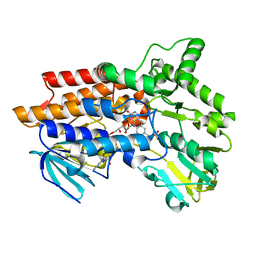 | | Phenylacetone Monooxygenase, a Baeyer-Villiger Monooxygenase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PHENYLACETONE MONOOXYGENASE, SULFATE ION | | Authors: | Malito, E, Alfieri, A, Mattevi, A. | | Deposit date: | 2004-08-03 | | Release date: | 2004-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of a Baeyer-Villiger Monooxygenase
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
2Y7S
 
 | |
4BIH
 
 | | Crystal structure of the conserved staphylococcal antigen 1A, Csa1A | | Descriptor: | CALCIUM ION, UNCHARACTERIZED LIPOPROTEIN SAOUHSC_00053 | | Authors: | Malito, E, Bottomley, M.J, Spraggon, G, Schluepen, C, Liberatori, S. | | Deposit date: | 2013-04-10 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.459 Å) | | Cite: | Mining the Bacterial Unknown Proteome: Identification and Characterization of a Novel Family of Highly Conserved Protective Antigens in Staphylococcus Aureus
Biochem.J., 455, 2013
|
|
4CJD
 
 | | Crystal structure of Neisseria meningitidis trimeric autotransporter and vaccine antigen NadA | | Descriptor: | IODIDE ION, NADA | | Authors: | Malito, E, Biancucci, M, Spraggon, G, Bottomley, M.J. | | Deposit date: | 2013-12-19 | | Release date: | 2014-11-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.056 Å) | | Cite: | Structure of the Meningococcal Vaccine Antigen Nada and Epitope Mapping of a Bactericidal Antibody.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4BIG
 
 | | Crystal structure of the conserved staphylococcal antigen 1B, Csa1B | | Descriptor: | UNCHARACTERIZED LIPOPROTEIN SAOUHSC_00053 | | Authors: | Malito, E, Bottomley, M.J, Schluepen, C, Liberatori, S. | | Deposit date: | 2013-04-10 | | Release date: | 2013-08-07 | | Last modified: | 2013-10-30 | | Method: | X-RAY DIFFRACTION (2.274 Å) | | Cite: | Mining the Bacterial Unknown Proteome: Identification and Characterization of a Novel Family of Highly Conserved Protective Antigens in Staphylococcus Aureus
Biochem.J., 455, 2013
|
|
4B8Y
 
 | | Ferrichrome-bound FhuD2 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FE (III) ION, ... | | Authors: | Malito, E, Bottomley, M.J, Spraggon, G. | | Deposit date: | 2012-08-31 | | Release date: | 2012-11-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Characterization of the Staphylococcus Aureus Virulence Factor and Vaccine Candidate Fhud2.
Biochem.J., 449, 2013
|
|
4D7W
 
 | | Crystal structure of sortase C1 (SrtC1) from Streptococcus agalactiae | | Descriptor: | 1,2-ETHANEDIOL, SORTASE FAMILY PROTEIN | | Authors: | Malito, E, Lazzarin, M, Cozzi, R, Bottomley, M.J. | | Deposit date: | 2014-11-28 | | Release date: | 2015-08-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Noncanonical Sortase-Mediated Assembly of Pilus Type 2B in Group B Streptococcus.
Faseb J., 29, 2015
|
|
4DMW
 
 | | Crystal structure of the GT domain of Clostridium difficile toxin A (TcdA) in complex with UDP and Manganese | | Descriptor: | MANGANESE (II) ION, Toxin A, URIDINE-5'-DIPHOSPHATE | | Authors: | Malito, E, D'Urzo, N, Bottomley, M.J, Biancucci, M, Scarselli, M, Maione, D, Martinelli, M. | | Deposit date: | 2012-02-08 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of Clostridium difficile toxin A glucosyltransferase domain bound to Mn2+ and UDP provides insights into glucosyltransferase activity and product release.
Febs J., 279, 2012
|
|
4DMV
 
 | | Crystal structure of the GT domain of Clostridium difficile Toxin A | | Descriptor: | Toxin A | | Authors: | Malito, E, D'Urzo, N, Bottomley, M.J, Biancucci, M, Maione, D, Scarselli, M, Martinelli, M. | | Deposit date: | 2012-02-08 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of Clostridium difficile toxin A glucosyltransferase domain bound to Mn2+ and UDP provides insights into glucosyltransferase activity and product release.
Febs J., 279, 2012
|
|
7SBZ
 
 | |
2VQ7
 
 | | Bacterial flavin-containing monooxygenase in complex with NADP: native data | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Alfieri, A, Malito, E, Orru, R, Fraaije, M.W, Mattevi, A. | | Deposit date: | 2008-03-12 | | Release date: | 2008-04-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Revealing the Moonlighting Role of Nadp in the Structure of a Flavin-Containing Monooxygenase.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
7MMN
 
 | |
7SA6
 
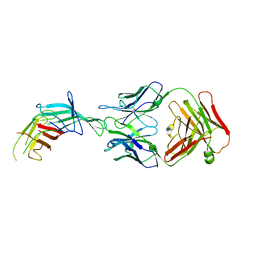 | | fHbp mutant 2416 bound to Fab JAR5 | | Descriptor: | Factor H-binding protein 2416, JAR5 Heavy Chain, JAR5 Light Chain | | Authors: | Chesterman, C, Malito, E, Bottomley, M.J. | | Deposit date: | 2021-09-22 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Active Learning for Rapid Design: An iterative AI approach for accelerated vaccine design that combines active machine learning and high-throughput experimental evaluation
To Be Published
|
|
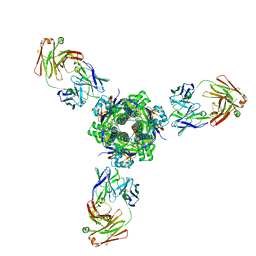
7MPG
 
 | |
7NRU
 
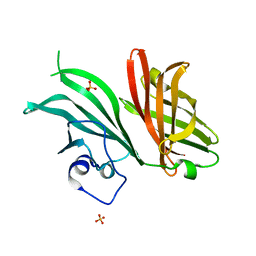 | |
5AIQ
 
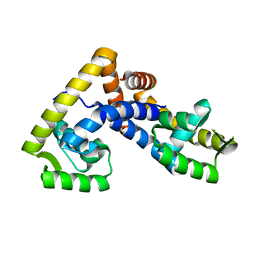 | | Crystal structure of ligand-free NadR | | Descriptor: | TRANSCRIPTIONAL REGULATOR, MARR FAMILY | | Authors: | Liguori, A, Malito, E, Bottomley, M.J. | | Deposit date: | 2015-02-16 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.716 Å) | | Cite: | Molecular Basis of Ligand-Dependent Regulation of Nadr, the Transcriptional Repressor of Meningococcal Virulence Factor Nada.
Plos Pathog., 12, 2016
|
|
5AIP
 
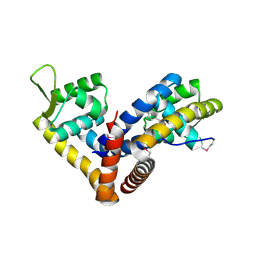 | | Crystal structure of NadR in complex with 4-hydroxyphenylacetate | | Descriptor: | 4-HYDROXYPHENYLACETATE, TRANSCRIPTIONAL REGULATOR, MARR FAMILY | | Authors: | Liguori, A, Malito, E, Bottomley, M.J. | | Deposit date: | 2015-02-16 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular Basis of Ligand-Dependent Regulation of Nadr, the Transcriptional Repressor of Meningococcal Virulence Factor Nada.
Plos Pathog., 12, 2016
|
|
6EUN
 
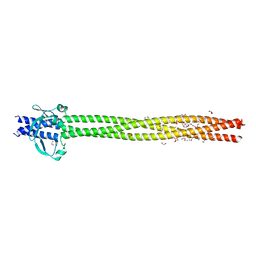 | | Crystal structure of Neisseria meningitidis vaccine antigen NadA variant 3 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, ... | | Authors: | Dello Iacono, L, Liguori, A, Malito, E, Bottomley, M.J. | | Deposit date: | 2017-10-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | NadA3 Structures Reveal Undecad Coiled Coils and LOX1 Binding Regions Competed by Meningococcus B Vaccine-Elicited Human Antibodies.
MBio, 9, 2018
|
|
6EUP
 
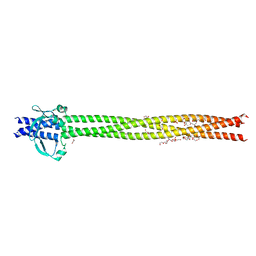 | | Crystal structure of Neisseria meningitidis NadA variant 3 double mutant A33I-I38L | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, Adhesin A, ... | | Authors: | Dello Iacono, L, Liguori, A, Malito, E, Bottomley, M.J. | | Deposit date: | 2017-10-31 | | Release date: | 2018-10-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | NadA3 Structures Reveal Undecad Coiled Coils and LOX1 Binding Regions Competed by Meningococcus B Vaccine-Elicited Human Antibodies.
MBio, 9, 2018
|
|
4OXR
 
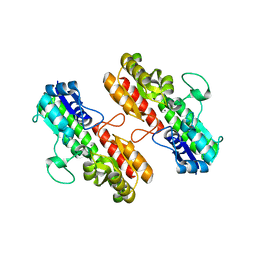 | | Structure of Staphylococcus pseudintermedius metal-binding protein SitA in complex with Manganese | | Descriptor: | MANGANESE (II) ION, Manganese ABC transporter, periplasmic-binding protein SitA | | Authors: | Abate, F, Malito, E, Bottomley, M. | | Deposit date: | 2014-02-06 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Apo, Zn2+-bound and Mn2+-bound structures reveal ligand-binding properties of SitA from the pathogen Staphylococcus pseudintermedius.
Biosci.Rep., 34, 2014
|
|
4OXQ
 
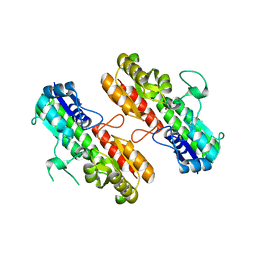 | | Structure of Staphylococcus pseudintermedius metal-binding protein SitA in complex with Zinc | | Descriptor: | Manganese ABC transporter, periplasmic-binding protein SitA, ZINC ION | | Authors: | Abate, F, Malito, E, Bottomley, M. | | Deposit date: | 2014-02-06 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Apo, Zn2+-bound and Mn2+-bound structures reveal ligand-binding properties of SitA from the pathogen Staphylococcus pseudintermedius.
Biosci.Rep., 34, 2014
|
|
3CQF
 
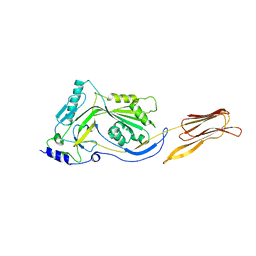 | | Crystal structure of anthrolysin O (ALO) | | Descriptor: | Thiol-activated cytolysin | | Authors: | Bourdeau, R.W, Malito, E, Tang, W.J. | | Deposit date: | 2008-04-02 | | Release date: | 2009-03-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Cellular Functions and X-ray Structure of Anthrolysin O, a Cholesterol-dependent Cytolysin Secreted by Bacillus anthracis
J.Biol.Chem., 284, 2009
|
|
5N4G
 
 | |
