2D6C
 
 | | Crystal structure of myoglobin reconstituted with iron porphycene | | Descriptor: | IMIDAZOLE, Myoglobin, PORPHYCENE CONTAINING FE | | Authors: | Hayashi, T, Murata, D, Makino, M, Sugimoto, H, Matsuo, T, Sato, H, Shiro, Y, Hisaeda, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-11-11 | | Release date: | 2006-10-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure and peroxidase activity of myoglobin reconstituted with iron porphycene
Inorg.Chem., 45, 2006
|
|
6EW2
 
 | | Human myelin protein P2 F57A mutant, tetragonal crystal form | | Descriptor: | CHLORIDE ION, Myelin P2 protein, PALMITIC ACID | | Authors: | Laulumaa, S, Lehtimaki, M, Kursula, P. | | Deposit date: | 2017-11-03 | | Release date: | 2018-07-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.588 Å) | | Cite: | Structure and dynamics of a human myelin protein P2 portal region mutant indicate opening of the beta barrel in fatty acid binding proteins.
BMC Struct. Biol., 18, 2018
|
|
6EW5
 
 | |
5NE0
 
 | | Room temperature in-situ structure of hen egg-white lysozyme from crystals enclosed between ultrathin silicon nitride membranes | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Martiel, I, Opara, N, Arnold, S.A, Braun, T, Stahlberg, H, Makita, M, David, C, Padeste, C. | | Deposit date: | 2017-03-09 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Direct protein crystallization on ultrathin membranes for diffraction measurements at X-ray free-electron lasers.
J.Appl.Crystallogr., 50, 2017
|
|
6STS
 
 | | Human myelin protein P2 mutant R30Q | | Descriptor: | Myelin P2 protein, PALMITIC ACID, SULFATE ION | | Authors: | Ruskamo, S, Lehtimaki, M, Kursula, P. | | Deposit date: | 2019-09-11 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cryo-EM, X-ray diffraction, and atomistic simulations reveal determinants for the formation of a supramolecular myelin-like proteolipid lattice.
J.Biol.Chem., 295, 2020
|
|
1PDU
 
 | | Ligand-binding domain of Drosophila orphan nuclear receptor DHR38 | | Descriptor: | nuclear hormone receptor HR38 | | Authors: | Baker, K.D, Shewchuk, L.M, Korlova, T, Makishima, M, Hassell, A.M, Wisely, B, Caravella, J.A, Lambert, M.H, Wilson, T.M, Mangelsdorf, D.J. | | Deposit date: | 2003-05-20 | | Release date: | 2003-06-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Drosophila orphan nuclear receptor DHR38 mediates an atypical ecdysteroid signaling pathway.
Cell(Cambridge,Mass.), 113, 2003
|
|
5XZH
 
 | | Vitamin D receptor with a synthetic ligand ADRO2 | | Descriptor: | (1R,3S,5Z)-5-[(2E)-2-[(1R,3aS,7aR)-1-[(2R,6R)-6-(1-adamantyl)-6-oxidanyl-hex-4-yn-2-yl]-7a-methyl-2,3,3a,5,6,7-hexahydro-1H-inden-4-ylidene]ethylidene]-4-methylidene-cyclohexane-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Otero, R, Numoto, N, Ikura, T, Yamada, S, Mourino, A, Makishima, M, Ito, N. | | Deposit date: | 2017-07-12 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 25 S-Adamantyl-23-yne-26,27-dinor-1 alpha ,25-dihydroxyvitamin D3: Synthesis, Tissue Selective Biological Activities, and X-ray Crystal Structural Analysis of Its Vitamin D Receptor Complex.
J. Med. Chem., 61, 2018
|
|
7CFO
 
 | | Crystal structure of human RXRalpha ligand binding domain complexed with CBTF-EE. | | Descriptor: | 1-[3-(2-ethoxyethoxy)-5,5,8,8-tetramethyl-6,7-dihydronaphthalen-2-yl]-2-(trifluoromethyl)benzimidazole-5-carboxylic acid, GLYCEROL, Retinoic acid receptor RXR-alpha | | Authors: | Watanabe, M, Fujihara, M, Motoyama, T, Kawasaki, M, Yamada, S, Takamura, Y, Ito, S, Makishima, M, Nakano, S, Kakuta, H. | | Deposit date: | 2020-06-27 | | Release date: | 2021-01-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of a "Gatekeeper" Antagonist that Blocks Entry Pathway to Retinoid X Receptors (RXRs) without Allosteric Ligand Inhibition in Permissive RXR Heterodimers.
J.Med.Chem., 64, 2021
|
|
5XZF
 
 | | Vitamin D receptor with a synthetic ligand ADRO1 | | Descriptor: | (1R,3S,5Z)-5-[(2E)-2-[(1R,3aS,7aR)-1-[(2R,6S)-6-(1-adamantyl)-6-oxidanyl-hex-4-yn-2-yl]-7a-methyl-2,3,3a,5,6,7-hexahydro-1H-inden-4-ylidene]ethylidene]-4-methylidene-cyclohexane-1,3-diol, FORMIC ACID, Mediator of RNA polymerase II transcription subunit 1, ... | | Authors: | Otero, R, Numoto, N, Ikura, T, Yamada, S, Mourino, A, Makishima, M, Ito, N. | | Deposit date: | 2017-07-12 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 25 S-Adamantyl-23-yne-26,27-dinor-1 alpha ,25-dihydroxyvitamin D3: Synthesis, Tissue Selective Biological Activities, and X-ray Crystal Structural Analysis of Its Vitamin D Receptor Complex.
J. Med. Chem., 61, 2018
|
|
6XVY
 
 | | Human myelin protein P2 mutant R88Q | | Descriptor: | CHLORIDE ION, CITRIC ACID, Myelin P2 protein, ... | | Authors: | Ruskamo, S, Lehtimaki, M, Kursula, P. | | Deposit date: | 2020-01-22 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cryo-EM, X-ray diffraction, and atomistic simulations reveal determinants for the formation of a supramolecular myelin-like proteolipid lattice.
J.Biol.Chem., 295, 2020
|
|
2EKT
 
 | | Crystal structure of myoglobin reconstituted with 6-methyl-6-depropionatehemin | | Descriptor: | 6-METHY-6-DEPROPIONATEHEMIN, Myoglobin, SULFATE ION | | Authors: | Harada, K, Makino, M, Sugimoto, H, Hirota, S, Matsuo, T, Shiro, Y, Hisaeda, Y, Hayashi, T. | | Deposit date: | 2007-03-25 | | Release date: | 2007-08-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure and ligand binding properties of myoglobins reconstituted with monodepropionated heme: functional role of each heme propionate side chain
Biochemistry, 46, 2007
|
|
6XUW
 
 | | Human myelin protein P2 mutant L27D | | Descriptor: | CHLORIDE ION, Myelin P2 protein, PALMITIC ACID | | Authors: | Ruskamo, S, Lehtimaki, M, Kursula, P. | | Deposit date: | 2020-01-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Cryo-EM, X-ray diffraction, and atomistic simulations reveal determinants for the formation of a supramolecular myelin-like proteolipid lattice.
J.Biol.Chem., 295, 2020
|
|
6XVQ
 
 | | Human myelin protein P2 mutant K31Q | | Descriptor: | CITRIC ACID, Myelin P2 protein, PALMITIC ACID | | Authors: | Ruskamo, S, Lehtimaki, M, Kursula, P. | | Deposit date: | 2020-01-22 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cryo-EM, X-ray diffraction, and atomistic simulations reveal determinants for the formation of a supramolecular myelin-like proteolipid lattice.
J.Biol.Chem., 295, 2020
|
|
6XVR
 
 | | Human myelin protein P2 mutant L35S | | Descriptor: | Myelin P2 protein, PALMITIC ACID, SULFATE ION | | Authors: | Ruskamo, S, Lehtimaki, M, Kursula, P. | | Deposit date: | 2020-01-22 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cryo-EM, X-ray diffraction, and atomistic simulations reveal determinants for the formation of a supramolecular myelin-like proteolipid lattice.
J.Biol.Chem., 295, 2020
|
|
2EKU
 
 | | Crystal structure of myoglobin reconstituted with 7-methyl-7-depropionatehemin | | Descriptor: | 7-METHYL-7-DEPROPIONATEHEMIN, Myoglobin, SULFATE ION | | Authors: | Harada, K, Makino, M, Sugimoto, H, Hirota, S, Matsuo, T, Shiro, Y, Hisaeda, Y, Hayashi, T. | | Deposit date: | 2007-03-25 | | Release date: | 2007-08-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and ligand binding properties of myoglobins reconstituted with monodepropionated heme: functional role of each heme propionate side chain
Biochemistry, 46, 2007
|
|
6XU9
 
 | | Human myelin protein P2 mutant K3N | | Descriptor: | CHLORIDE ION, CITRIC ACID, Myelin P2 protein, ... | | Authors: | Ruskamo, S, Lehtimaki, M, Kursula, P. | | Deposit date: | 2020-01-17 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cryo-EM, X-ray diffraction, and atomistic simulations reveal determinants for the formation of a supramolecular myelin-like proteolipid lattice.
J.Biol.Chem., 295, 2020
|
|
6XVS
 
 | | Human myelin protein P2 mutant P38G, unliganded | | Descriptor: | GLYCEROL, Myelin P2 protein | | Authors: | Ruskamo, S, Lehtimaki, M, Kursula, P. | | Deposit date: | 2020-01-22 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cryo-EM, X-ray diffraction, and atomistic simulations reveal determinants for the formation of a supramolecular myelin-like proteolipid lattice.
J.Biol.Chem., 295, 2020
|
|
6XW9
 
 | | Human myelin protein P2 mutant K120S | | Descriptor: | CHLORIDE ION, Myelin P2 protein, PALMITIC ACID | | Authors: | Ruskamo, S, Lehtimaki, M, Kursula, P. | | Deposit date: | 2020-01-23 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Cryo-EM, X-ray diffraction, and atomistic simulations reveal determinants for the formation of a supramolecular myelin-like proteolipid lattice.
J.Biol.Chem., 295, 2020
|
|
6XUA
 
 | | Human myelin protein P2 mutant K21Q | | Descriptor: | CITRIC ACID, Myelin P2 protein, PALMITIC ACID | | Authors: | Ruskamo, S, Lehtimaki, M, Kursula, P. | | Deposit date: | 2020-01-17 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cryo-EM, X-ray diffraction, and atomistic simulations reveal determinants for the formation of a supramolecular myelin-like proteolipid lattice.
J.Biol.Chem., 295, 2020
|
|
6XU5
 
 | | Human myelin protein P2 mutant N2D | | Descriptor: | CITRIC ACID, Myelin P2 protein, PALMITIC ACID | | Authors: | Ruskamo, S, Lehtimaki, M, Kursula, P. | | Deposit date: | 2020-01-17 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Cryo-EM, X-ray diffraction, and atomistic simulations reveal determinants for the formation of a supramolecular myelin-like proteolipid lattice.
J.Biol.Chem., 295, 2020
|
|
1WP1
 
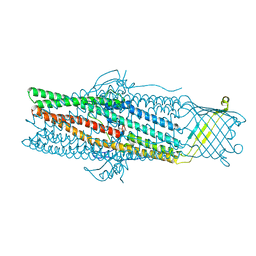 | | Crystal structure of the drug-discharge outer membrane protein, OprM | | Descriptor: | Outer membrane protein oprM | | Authors: | Akama, H, Kanemaki, M, Yoshimura, M, Tsukihara, T, Kashiwagi, T, Narita, S, Nakagawa, A, Nakae, T. | | Deposit date: | 2004-08-28 | | Release date: | 2004-11-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Crystal structure of the drug discharge outer membrane protein, OprM, of Pseudomonas aeruginosa: dual modes of membrane anchoring and occluded cavity end
J.Biol.Chem., 279, 2004
|
|
2YQ9
 
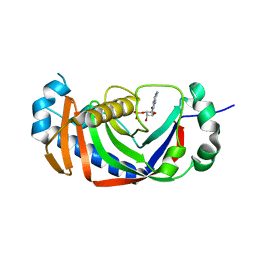 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, with mutation V321A, crystallized with 2'-AMP | | Descriptor: | 2', 3'-CYCLIC-NUCLEOTIDE 3'-PHOSPHODIESTERASE, ADENOSINE-2'-MONOPHOSPHATE | | Authors: | Myllykoski, M, Raasakka, A, Lehtimaki, M, Han, H, Kursula, P. | | Deposit date: | 2012-11-07 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic Analysis of the Reaction Cycle of 2',3'-Cyclic Nucleotide 3'-Phosphodiesterase, a Unique Member of the 2H Phosphoesterase Family
J.Mol.Biol., 425, 2013
|
|
2YPH
 
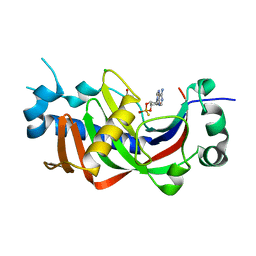 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, with mutation H230S, crystallized with 2',3-(RP)- cyclic-AMPS | | Descriptor: | 2', 3'-CYCLIC-NUCLEOTIDE 3'-PHOSPHODIESTERASE, ACETATE ION, ... | | Authors: | Myllykoski, M, Raasakka, A, Lehtimaki, M, Han, H, Kursula, P. | | Deposit date: | 2012-10-30 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic Analysis of the Reaction Cycle of 2',3'-Cyclic Nucleotide 3'-Phosphodiesterase, a Unique Member of the 2H Phosphoesterase Family
J.Mol.Biol., 425, 2013
|
|
2ZMH
 
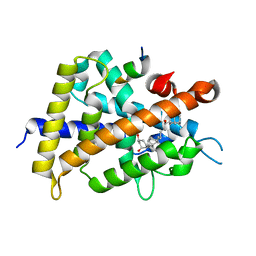 | | Crystal Structure of Rat Vitamin D Receptor Bound to Adamantyl Vitamin D Analogs: Structural Basis for Vitamin D Receptor Antagonism and/or Partial Agonism | | Descriptor: | (1R,3R,7E,17beta)-17-{(1R,2E,4R)-4-hydroxy-1-methyl-4-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]dec-1-yl]but-2-en-1-yl}-2-methylidene-9,10-secoestra-5,7-diene-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Nakabayashi, M, Yamada, S, Tanaka, T, Igarashi, M, Yoshimoto, N, Ikura, T, Ito, N, Makishima, M, Tokiwa, H, DeLuca, H.F, Shimizu, M. | | Deposit date: | 2008-04-18 | | Release date: | 2008-09-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of rat vitamin d receptor bound to adamantyl vitamin d analogs: structural basis for vitamin d receptor antagonism and partial agonism
J.Med.Chem., 51, 2008
|
|
1V5H
 
 | | Crystal Structure of Human Cytoglobin (Ferric Form) | | Descriptor: | Cytoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugimoto, H, Makino, M, Sawai, H, Kawada, N, Yoshizato, K, Shiro, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-23 | | Release date: | 2004-06-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of human cytoglobin for ligand binding.
J.Mol.Biol., 339, 2004
|
|
