3WRU
 
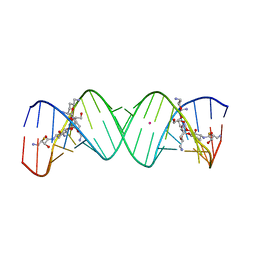 | | Crystal structure of the bacterial ribosomal decoding site in complex with synthetic aminoglycoside with F-HABA group | | Descriptor: | (2R,3R)-4-amino-N-[(1R,2S,3R,4R,5S)-5-amino-4-[(2,6-diamino-2,3,4,6-tetradeoxy-alpha-D-erythro-hexopyranosyl)oxy]-3-{[3-O-(2,6-diamino-2,3,4,6-tetradeoxy-beta-L-threo-hexopyranosyl)-beta-D-ribofuranosyl]oxy}-2-hydroxycyclohexyl]-3-fluoro-2-hydroxybutanamide, POTASSIUM ION, RNA (5'-R(*UP*UP*GP*CP*GP*UP*CP*AP*CP*GP*CP*CP*GP*GP*CP*GP*AP*AP*GP*UP*CP*GP*C)-3') | | Authors: | Maianti, J.P, Kanazawa, H, Dozzo, P, Feeney, L.A, Armstrong, E.S, Kondo, J, Hanessian, S. | | Deposit date: | 2014-02-27 | | Release date: | 2014-11-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Toxicity Modulation, Resistance Enzyme Evasion, and A-Site X-ray Structure of Broad-Spectrum Antibacterial Neomycin Analogs
Acs Chem.Biol., 9, 2014
|
|
6EDS
 
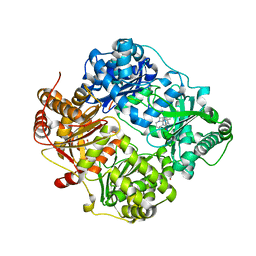 | | Structure of Cysteine-free Human Insulin-Degrading Enzyme in complex with Glucagon and Substrate-selective Macrocyclic Inhibitor 63 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tan, G.A, Seeliger, M.A, Maianti, J.P, Liu, D.R, Welsh, A.J. | | Deposit date: | 2018-08-10 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.18071723 Å) | | Cite: | Substrate-selective inhibitors that reprogram the activity of insulin-degrading enzyme.
Nat.Chem.Biol., 15, 2019
|
|
6MQ3
 
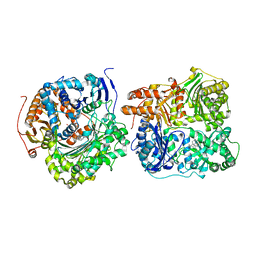 | | Structure of Cysteine-free Human Insulin-Degrading Enzyme in complex with Substrate-selective Macrocycle Inhibitor 63 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Insulin-degrading enzyme, {(8R,9S,10S)-9-(2',3'-dimethyl[1,1'-biphenyl]-4-yl)-6-[(1-methyl-1H-imidazol-2-yl)sulfonyl]-1,6-diazabicyclo[6.2.0]decan-10-yl}methanol | | Authors: | Tan, G.A, Seeliger, M.A, Welsh, A.J, Maianti, J.P, Liu, D.R. | | Deposit date: | 2018-10-09 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.569147 Å) | | Cite: | Substrate-selective inhibitors that reprogram the activity of insulin-degrading enzyme.
Nat.Chem.Biol., 15, 2019
|
|
6BYZ
 
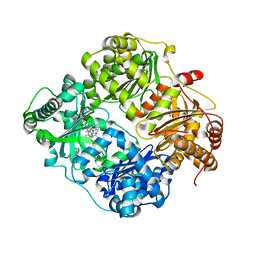 | | Structure of Cysteine-free Human Insulin-Degrading Enzyme in complex with Substrate-selective Macrocyclic Inhibitor 37 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ALA-ALA-ALA, Insulin-degrading enzyme, ... | | Authors: | Tan, G.A, Seeliger, M.A, Maianti, J.P, Liu, D.R. | | Deposit date: | 2017-12-21 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.95624852 Å) | | Cite: | Substrate-selective inhibitors that reprogram the activity of insulin-degrading enzyme.
Nat.Chem.Biol., 15, 2019
|
|
4PDQ
 
 | | Crystal structure of the bacterial ribosomal decoding site in complex with 4'-deoxy-4'-fluoro neomycin analog | | Descriptor: | (2S)-4-amino-N-{(1R,2S,3R,4R,5S)-5-amino-3-{[3-O-(2,6-diamino-2,6-dideoxy-beta-L-idopyranosyl)-beta-D-ribofuranosyl]oxy }-4-[(2,6-diamino-2,4,6-trideoxy-4-fluoro-alpha-D-galactopyranosyl)oxy]-2-hydroxycyclohexyl}-2-hydroxybutanamide, MAGNESIUM ION, RNA (5'-*UP*UP*GP*CP*GP*UP*CP*AP*CP*GP*CP*CP*GP*GP*CP*GP*AP*AP*GP*UP*CP*GP*C-3') | | Authors: | Hanessian, S, Saavedra, O.M, Vilchis-Reyes, M.A, Maianti, J.P, Kanazawa, H, Dozzo, P, Feeney, L.A, Armstrong, E.S, Kondo, J. | | Deposit date: | 2014-04-21 | | Release date: | 2015-01-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Synthesis, broad spectrum antibacterial activity, and X-ray co-crystal structure of the decoding bacterial ribosomal A-site with 4'-deoxy-4'-fluoro neomycin analogs
Chem Sci, 5, 2014
|
|
4GPX
 
 | | Crystal structure of the protozoal cytoplasmic ribosomal decoding site in complex with 6'-hydroxysisomicin (P212121 form) | | Descriptor: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(hydroxymethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, RNA (5'-R(*UP*UP*GP*CP*GP*UP*CP*GP*CP*GP*CP*CP*GP*GP*CP*GP*AP*AP*GP*UP*CP*GP*C)-3') | | Authors: | Kondo, J, Koganei, M, Maianti, J.P, Ly, V.L, Hanessian, S. | | Deposit date: | 2012-08-22 | | Release date: | 2013-04-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of a bioactive 6'-hydroxy variant of sisomicin bound to the bacterial and protozoal ribosomal decoding sites
Chemmedchem, 8, 2013
|
|
4GPW
 
 | | Crystal structure of the protozoal cytoplasmic ribosomal decoding site in complex with 6'-hydroxysisomicin (P21212 form) | | Descriptor: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(hydroxymethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, RNA (5'-R(*UP*UP*GP*CP*GP*UP*CP*GP*CP*GP*CP*CP*GP*GP*CP*GP*AP*AP*GP*UP*CP*GP*C)-3') | | Authors: | Kondo, J, Koganei, M, Maianti, J.P, Ly, V.L, Hanessian, S. | | Deposit date: | 2012-08-22 | | Release date: | 2013-04-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of a bioactive 6'-hydroxy variant of sisomicin bound to the bacterial and protozoal ribosomal decoding sites
Chemmedchem, 8, 2013
|
|
4GPY
 
 | | Crystal structure of the bacterial ribosomal decoding site in complex with 6'-hydroxysisomicin | | Descriptor: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(hydroxymethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, RNA (5'-R(*UP*UP*GP*CP*GP*UP*CP*AP*CP*GP*CP*CP*GP*GP*CP*GP*AP*AP*GP*UP*CP*GP*C)-3') | | Authors: | Kondo, J, Koganei, M, Maianti, J.P, Ly, V.L, Hanessian, S. | | Deposit date: | 2012-08-22 | | Release date: | 2013-04-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of a bioactive 6'-hydroxy variant of sisomicin bound to the bacterial and protozoal ribosomal decoding sites
Chemmedchem, 8, 2013
|
|
