8RZ3
 
 | | Structures of Se- glycosyltransferase SenB from Variovorax paradoxus | | Descriptor: | TIGR04348 family glycosyltransferase, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Ma, Y.Y, Gao, Y, Xu, S.H. | | Deposit date: | 2024-02-12 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structures of SenB and SenA enzymes from Variovorax paradoxus provide insights into carbon-selenium bond formation in selenoneine biosynthesis.
Heliyon, 10, 2024
|
|
8RYZ
 
 | | Structures of selenoneine synthase SenA from Variovorax paradoxus | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, IMIDAZOLE, ... | | Authors: | Ma, Y.Y, Gao, Y, Xu, S.H. | | Deposit date: | 2024-02-11 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structures of SenB and SenA enzymes from Variovorax paradoxus provide insights into carbon-selenium bond formation in selenoneine biosynthesis.
Heliyon, 10, 2024
|
|
5C8S
 
 | | Crystal structure of the SARS coronavirus nsp14-nsp10 complex with functional ligands SAH and GpppA | | Descriptor: | GUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, Guanine-N7 methyltransferase, MAGNESIUM ION, ... | | Authors: | Ma, Y.Y, Wu, L.J, Zhang, R.G, Rao, Z.H. | | Deposit date: | 2015-06-26 | | Release date: | 2015-07-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.326 Å) | | Cite: | Structural basis and functional analysis of the SARS coronavirus nsp14-nsp10 complex
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5C8T
 
 | | Crystal structure of the SARS coronavirus nsp14-nsp10 complex with functional ligand SAM | | Descriptor: | Guanine-N7 methyltransferase, MAGNESIUM ION, Non-structural protein 10, ... | | Authors: | Ma, Y.Y, Wu, L.J, Zhang, R.G, Rao, Z.H. | | Deposit date: | 2015-06-26 | | Release date: | 2015-07-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis and functional analysis of the SARS coronavirus nsp14-nsp10 complex
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5C8U
 
 | | Crystal structure of the SARS coronavirus nsp14-nsp10 complex | | Descriptor: | Guanine-N7 methyltransferase, MAGNESIUM ION, Non-structural protein 10, ... | | Authors: | Ma, Y.Y, Wu, L.J, Zhang, R.G, Rao, Z.H. | | Deposit date: | 2015-06-26 | | Release date: | 2015-07-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.401 Å) | | Cite: | Structural basis and functional analysis of the SARS coronavirus nsp14-nsp10 complex
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3UIM
 
 | |
3T35
 
 | | Arabidopsis thaliana dynamin-related protein 1A in postfission state | | Descriptor: | Dynamin-related protein 1A, LINKER, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Yan, L.M, Ma, Y.Y, Sun, Y.N, Lou, Z.Y. | | Deposit date: | 2011-07-24 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.592 Å) | | Cite: | Structural basis for mechanochemical role of Arabidopsis thaliana dynamin-related protein in membrane fission
J Mol Cell Biol, 3, 2011
|
|
3T34
 
 | | Arabidopsis thaliana dynamin-related protein 1A (AtDRP1A) in prefission state | | Descriptor: | Dynamin-related protein 1A, LINKER, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Yan, L.M, Ma, Y.Y, Sun, Y.N, Lou, Z.Y. | | Deposit date: | 2011-07-24 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.405 Å) | | Cite: | Structural basis for mechanochemical role of Arabidopsis thaliana dynamin-related protein in membrane fission
J Mol Cell Biol, 3, 2011
|
|
3ULZ
 
 | | Crystal structure of apo BAK1 | | Descriptor: | BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 | | Authors: | Lou, Z.Y, Yan, L.M, Ma, Y.Y. | | Deposit date: | 2011-11-11 | | Release date: | 2012-11-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for BAK1 activation
To be Published
|
|
8IKS
 
 | |
8IKP
 
 | |
8IHU
 
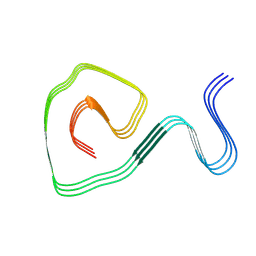 | | Cryo-EM structure of an amyloid fibril formed by ALS-causing SOD1 mutation G85R | | Descriptor: | Superoxide dismutase [Cu-Zn] | | Authors: | Wang, L.Q, Ma, Y.Y, Zhang, M.Y, Yuan, H.Y, Li, X.N, Zhao, K, Chen, J, Li, D, Wang, Z.Z, Le, W.D, Liu, C, Liang, Y. | | Deposit date: | 2023-02-23 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Amyloid fibril structures and ferroptosis activation induced by ALS-causing SOD1 mutations.
Sci Adv, 2024
|
|
8IHV
 
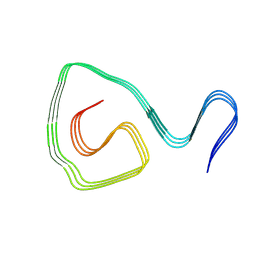 | | Cryo-EM structure of an amyloid fibril formed by ALS-causing SOD1 mutation H46R | | Descriptor: | Superoxide dismutase [Cu-Zn] | | Authors: | Wang, L.Q, Ma, Y.Y, Zhang, M.Y, Yuan, H.Y, Li, X.N, Zhao, K, Chen, J, Li, D, Wang, Z.Z, Le, W.D, Liu, C, Liang, Y. | | Deposit date: | 2023-02-23 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Amyloid fibril structures and ferroptosis activation induced by ALS-causing SOD1 mutations.
Sci Adv, 2024
|
|
8IY7
 
 | |
8IZZ
 
 | |
8IK7
 
 | |
8IKB
 
 | |
7DWV
 
 | | Cryo-EM structure of amyloid fibril formed by familial prion disease-related mutation E196K | | Descriptor: | Major prion protein | | Authors: | Wang, L.Q, Zhao, K, Yuan, H.Y, Li, X.N, Dang, H.B, Ma, Y.Y, Wang, Q, Wang, C, Sun, Y.P, Chen, J, Li, D, Zhang, D.L, Yin, P, Liu, C, Liang, Y. | | Deposit date: | 2021-01-18 | | Release date: | 2021-10-13 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Genetic prion disease-related mutation E196K displays a novel amyloid fibril structure revealed by cryo-EM.
Sci Adv, 7, 2021
|
|
7DA4
 
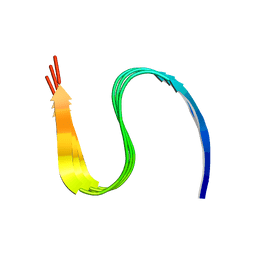 | | Cryo-EM structure of amyloid fibril formed by human RIPK3 | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 3 | | Authors: | Zhao, K, Ma, Y.Y, Sun, Y.P, Li, D, Liu, C. | | Deposit date: | 2020-10-14 | | Release date: | 2021-04-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.24 Å) | | Cite: | The structure of a minimum amyloid fibril core formed by necroptosis-mediating RHIM of human RIPK3.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7VZF
 
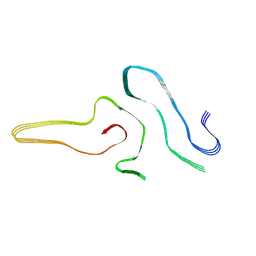 | | Cryo-EM structure of amyloid fibril formed by full-length human SOD1 | | Descriptor: | Superoxide dismutase [Cu-Zn] | | Authors: | Wang, L.Q, Ma, Y.Y, Yuan, H.Y, Zhao, K, Zhang, M.Y, Wang, Q, Huang, X, Xu, W.C, Chen, J, Li, D, Zhang, D.L, Zou, L.Y, Yin, P, Liu, C, Liang, Y. | | Deposit date: | 2021-11-16 | | Release date: | 2022-06-29 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Cryo-EM structure of an amyloid fibril formed by full-length human SOD1 reveals its conformational conversion.
Nat Commun, 13, 2022
|
|
