6U42
 
 | | Natively decorated ciliary doublet microtubule | | Descriptor: | DC1, DC2, DC3, ... | | Authors: | Ma, M, Stoyanova, M, Rademacher, G, Dutcher, S.K, Brown, A, Zhang, R. | | Deposit date: | 2019-08-22 | | Release date: | 2019-11-13 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the Decorated Ciliary Doublet Microtubule.
Cell, 179, 2019
|
|
7CU6
 
 | | lasso peptide C24 mutant - A11V2C | | Descriptor: | lasso peptide C24_A11V2C | | Authors: | Ma, M, Liu, X.H. | | Deposit date: | 2020-08-21 | | Release date: | 2021-08-25 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Rational generation of lasso peptides based on biosynthetic gene mutations and site-selective chemical modifications.
Chem Sci, 12, 2021
|
|
4Z5P
 
 | | Crystal structure of the LnmA cytochrome P450 hydroxylase from the leinamycin biosynthetic pathway of Streptomyces atroolivaceus S-140 at 1.9 A resolution | | Descriptor: | Cytochrome P450 hydroxylase, PROTOPORPHYRIN IX CONTAINING FE, TRIETHYLENE GLYCOL | | Authors: | Ma, M, Lohman, J, Rudolf, J, Miller, M.D, Cao, H, Osipiuk, J, Babnigg, G, Phillips Jr, G.N, Joachimiak, A, Shen, B, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-04-02 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the LnmA cytochrome P450 hydroxylase from the leinamycin biosynthetic pathway of Streptomyces atroolivaceus S-140
To be Published
|
|
4Z5Q
 
 | | Crystal structure of the LnmZ cytochrome P450 hydroxylase from the leinamycin biosynthetic pathway of Streptomyces atroolivaceus S-140 at 1.8 A resolution | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, Cytochrome P450 hydroxylase, ... | | Authors: | Ma, M, Lohman, J, Rudolf, J, Miller, M.D, Cao, H, Osipiuk, J, Joachimiak, A, Phillips Jr, G.N, Shen, B, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-04-02 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Crystal structure of the LnmZ cytochrome P450 hydroxylase from the leinamycin biosynthetic pathway of Streptomyces atroolivaceus S-140
To be Published
|
|
7XRF
 
 | | Crystal structaure of DgpB/C complex | | Descriptor: | AP_endonuc_2 domain-containing protein, DgpB, MANGANESE (II) ION | | Authors: | Ma, M, He, P. | | Deposit date: | 2022-05-10 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.137 Å) | | Cite: | Structural mechanism of a dual-functional enzyme DgpA/B/C as both a C -glycoside cleaving enzyme and an O - to C -glycoside isomerase.
Acta Pharm Sin B, 13, 2023
|
|
8YL9
 
 | | Crystal structures of terpene synthases complexed with a substrate mimic | | Descriptor: | MAGNESIUM ION, N-benzyl-N,N-diethylethanaminium, PYROPHOSPHATE, ... | | Authors: | Xu, M, Ma, M. | | Deposit date: | 2024-03-05 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Insights Into the Terpene Cyclization Domains of Two Fungal Sesterterpene Synthases and Enzymatic Engineering for Sesterterpene Diversification.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8YLA
 
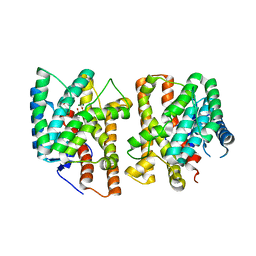 | | Crystal structures of terpene synthases complexed with a substrate mimic | | Descriptor: | MAGNESIUM ION, N-benzyl-N,N-diethylethanaminium, PYROPHOSPHATE, ... | | Authors: | Xu, M, Ma, M. | | Deposit date: | 2024-03-05 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural Insights Into the Terpene Cyclization Domains of Two Fungal Sesterterpene Synthases and Enzymatic Engineering for Sesterterpene Diversification.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
7XH2
 
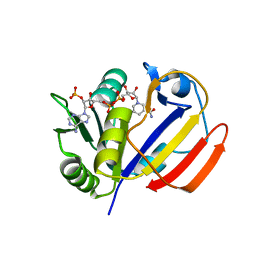 | | Dihydrofolate Reductase-like Protein SacH in safracin biosynthesis | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Uncharacterized protein sfcH | | Authors: | Ma, X, Shao, N, Ma, M, Tang, G. | | Deposit date: | 2022-04-07 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dihydrofolate reductase-like protein inactivates hemiaminal pharmacophore for self-resistance in safracin biosynthesis.
Acta Pharm Sin B, 13, 2023
|
|
7XH4
 
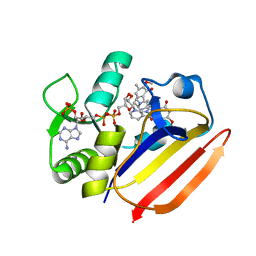 | | Dihydrofolate Reductase-like Protein SacH in safracin biosynthesis complex with safracin A | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Safracin A, Uncharacterized protein sfcH | | Authors: | Ma, X, Shao, N, Zhang, Y, Yang, D, Ma, M, Tang, G. | | Deposit date: | 2022-04-07 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dihydrofolate reductase-like protein inactivates hemiaminal pharmacophore for self-resistance in safracin biosynthesis.
Acta Pharm Sin B, 13, 2023
|
|
9B4H
 
 | | Chlamydomonas reinhardtii mastigoneme filament | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C-type lectin domain-containing protein, Tyrosine-protein kinase ephrin type A/B receptor-like domain-containing protein, ... | | Authors: | Dai, J, Ma, M, Zhang, R, Brown, A. | | Deposit date: | 2024-03-20 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mastigoneme structure reveals insights into the O-linked glycosylation code of native hydroxyproline-rich helices.
Cell, 2024
|
|
4WKY
 
 | | Streptomcyes albus JA3453 oxazolomycin ketosynthase domain OzmN KS2 | | Descriptor: | 1,2-ETHANEDIOL, Beta-ketoacyl synthase, GLYCEROL, ... | | Authors: | Cuff, M.E, Mack, J.C, Endres, M, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-10-03 | | Release date: | 2014-10-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6M19
 
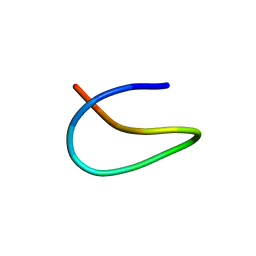 | | Template lasso peptide C24 mutant W14F | | Descriptor: | lasso peptide | | Authors: | Liu, X.H, Liu, T, Ma, X.J, Yu, J.H, Yang, D.H, Ma, M. | | Deposit date: | 2020-02-25 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rational generation of lasso peptides based on biosynthetic gene mutations and site-selective chemical modifications.
Chem Sci, 12, 2021
|
|
4TKT
 
 | | Streptomyces platensis isomigrastatin ketosynthase domain MgsF KS6 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, AT-less polyketide synthase, CHLORIDE ION, ... | | Authors: | Chang, C, Li, H, Endres, M, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-05-27 | | Release date: | 2014-06-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4289 Å) | | Cite: | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7BZA
 
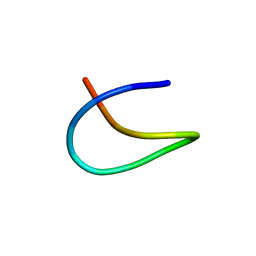 | | Template lasso peptide C24 | | Descriptor: | lasso peptide | | Authors: | Liu, X.H, Liu, T, Ma, X.J, Yu, J.H, Yang, D.H, Ma, M. | | Deposit date: | 2020-04-27 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rational generation of lasso peptides based on biosynthetic gene mutations and site-selective chemical modifications.
Chem Sci, 12, 2021
|
|
7BZ8
 
 | | Template lasso peptide C24 mutant V3A | | Descriptor: | lasso peptide | | Authors: | Liu, X.H, Liu, T, Ma, X.J, Yu, J.H, Yang, D.H, Ma, M. | | Deposit date: | 2020-04-27 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rational generation of lasso peptides based on biosynthetic gene mutations and site-selective chemical modifications.
Chem Sci, 12, 2021
|
|
7BZ9
 
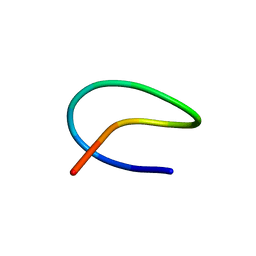 | | Template lasso peptide C24 mutant I4A | | Descriptor: | lasso peptide | | Authors: | Liu, X.H, Liu, T, Ma, X.J, Yu, J.H, Yang, D.H, Ma, M. | | Deposit date: | 2020-04-27 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rational generation of lasso peptides based on biosynthetic gene mutations and site-selective chemical modifications.
Chem Sci, 12, 2021
|
|
7XP7
 
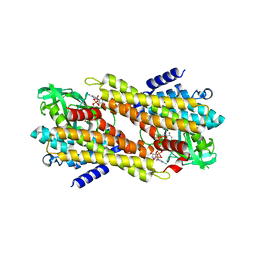 | | Crystal Structure of the Flavoprotein ColB1 Catalyzing Assembly Line-Tethered Cysteine Dehydrogenation | | Descriptor: | Cyclohexanecarboxyl-CoA dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Ma, X.Y, Tang, Z.J, Liu, W, Ma, M. | | Deposit date: | 2022-05-03 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Based Mechanistic Insights into ColB1, a Flavoprotein Functioning in-trans in the 2,2'-Bipyridine Assembly Line for Cysteine Dehydrogenation.
Acs Chem.Biol., 18, 2023
|
|
5BMO
 
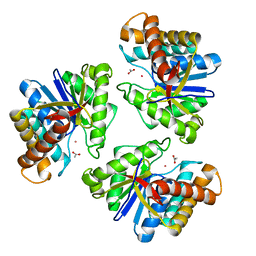 | | LnmX protein, a putative GlcNAc-PI de-N-acetylase from Streptomyces atroolivaceus | | Descriptor: | ACETATE ION, POTASSIUM ION, Putative uncharacterized protein LnmX | | Authors: | Osipiuk, J, Hatzos-Skintges, C, Cuff, M, Endres, M, Babnigg, G, Lohman, J, Ma, M, Rudolf, J, Chang, C.-Y, Shen, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-05-22 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | LnmX protein, a putative GlcNAc-PI de-N-acetylase from Streptomyces atroolivaceus.
to be published
|
|
4ZDN
 
 | | Streptomyces platensis isomigrastatin ketosynthase domain MgsF KS4 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AT-less polyketide synthase, CHLORIDE ION | | Authors: | Chang, C, Li, H, Endres, M, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-04-17 | | Release date: | 2015-05-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.509 Å) | | Cite: | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7BZ7
 
 | | Template lasso peptide C24 mutant F15Y | | Descriptor: | lasso peptide | | Authors: | Liu, X.H, Liu, T, Ma, X.J, Yu, J.H, Yang, D.H, Ma, M. | | Deposit date: | 2020-04-27 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rational generation of lasso peptides based on biosynthetic gene mutations and site-selective chemical modifications.
Chem Sci, 12, 2021
|
|
7DXO
 
 | |
7CRN
 
 | |
7Y50
 
 | | Class I diterpene synthase (CyS) from Streptomyces cattleya | | Descriptor: | Putative glutamate dehydrogenase/leucine dehydrogenase | | Authors: | Xing, B, Ma, M. | | Deposit date: | 2022-06-16 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure Based Mutagenesis of Cattleyene Synthase Leads to the Generation of Rearranged Polycyclic Diterpenes.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7Y87
 
 | |
7Y88
 
 | | class I diterpene synthase (CyS-GGPP-Mg2+) from Streptomyces cattleya | | Descriptor: | GERANYLGERANYL DIPHOSPHATE, MAGNESIUM ION, Putative glutamate dehydrogenase/leucine dehydrogenase | | Authors: | Xing, B, Ma, M. | | Deposit date: | 2022-06-23 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal Structure Based Mutagenesis of Cattleyene Synthase Leads to the Generation of Rearranged Polycyclic Diterpenes.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
