8XI5
 
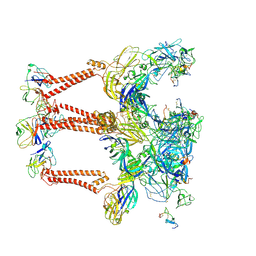 | | Structure of Eastern Equine Encephalitis VLP in complex with the receptor VLDLR LA3-5 | | Descriptor: | CALCIUM ION, Capsid protein, Spike glycoprotein E1, ... | | Authors: | Cao, D, Ma, B, Cao, Z, Xu, X, Zhang, X, Xiang, Y. | | Deposit date: | 2023-12-19 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The receptor VLDLR binds Eastern Equine Encephalitis virus through multiple distinct modes.
Nat Commun, 15, 2024
|
|
5F6X
 
 | | Crystal structure of Ubc9 (K48/K49A/E54A) complexed with Fragment 2 (mercaptobenzoxazole from cocktail screen) | | Descriptor: | 5-chloranyl-3~{H}-1,3-benzoxazole-2-thione, SUMO-conjugating enzyme UBC9 | | Authors: | Lountos, G.T, Hewitt, W.M, Zlotkowski, K, Dahlhauser, S, Saunders, L.B, Needle, D, Tropea, J.E, Zhan, C, Wei, G, Ma, B, Nussinov, R, Schneekloth, J.S.Jr, Waugh, D.S. | | Deposit date: | 2015-12-07 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Insights Into the Allosteric Inhibition of the SUMO E2 Enzyme Ubc9.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5F6Y
 
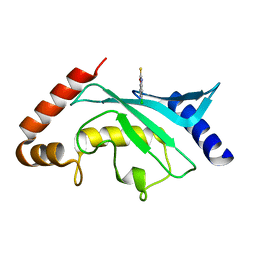 | | Crystal structure of Ubc9 (K48/K49A/E54A) complexed with Fragment 2 (mercaptobenzoxazole) | | Descriptor: | 5-chloranyl-3~{H}-1,3-benzoxazole-2-thione, SUMO-conjugating enzyme UBC9 | | Authors: | Lountos, G.T, Hewitt, W.M, Zlotkowski, K, Dahlhauser, S, Saunders, L.B, Needle, D, Tropea, J.E, Zhan, C, Wei, G, Ma, B, Nussinov, R, Schneekloth, J.S.Jr, Waugh, D.S. | | Deposit date: | 2015-12-07 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Insights Into the Allosteric Inhibition of the SUMO E2 Enzyme Ubc9.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
7UG2
 
 | | Crystal structure of the coiled-coil domain of TRIM75 | | Descriptor: | ACETYL GROUP, ISOPROPYL ALCOHOL, Tripartite motif-containing protein 75 | | Authors: | Lou, X.H, Ma, B.B, Zhuang, Y, Li, X.C. | | Deposit date: | 2022-03-23 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Structural studies of the coiled-coil domain of TRIM75 reveal a tetramer architecture facilitating its E3 ligase complex.
Comput Struct Biotechnol J, 20, 2022
|
|
7WCD
 
 | | Cryo EM structure of SARS-CoV-2 spike in complex with TAU-2212 mAbs in conformation 4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain, Light chain, ... | | Authors: | Xiang, Y, Ma, B, Li, R. | | Deposit date: | 2021-12-19 | | Release date: | 2022-08-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Conformational flexibility in neutralization of SARS-CoV-2 by naturally elicited anti-SARS-CoV-2 antibodies.
Commun Biol, 5, 2022
|
|
7WU7
 
 | | Prefoldin-tubulin-TRiC complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Prefoldin subunit 1, Prefoldin subunit 2, ... | | Authors: | Gestaut, D, Zhao, Y, Park, J, Ma, B, Leitner, A, Collier, M, Pintilie, G, Roh, S.-H, Chiu, W, Frydman, J. | | Deposit date: | 2022-02-07 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Structural visualization of the tubulin folding pathway directed by human chaperonin TRiC/CCT.
Cell, 185, 2022
|
|
7EVP
 
 | | Cryo-EM structure of the Gp168-beta-clamp complex | | Descriptor: | Beta sliding clamp, Sliding clamp inhibitor | | Authors: | Liu, B, Li, S, Liu, Y, Chen, H, Hu, Z, Wang, Z, Gou, L, Zhang, L, Ma, B, Wang, H, Matthews, S, Wang, Y, Zhang, K. | | Deposit date: | 2021-05-21 | | Release date: | 2022-02-16 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Bacteriophage Twort protein Gp168 is a beta-clamp inhibitor by occupying the DNA sliding channel.
Nucleic Acids Res., 49, 2021
|
|
6J7W
 
 | | Crystal Structure of Human BCMA in complex with UniAb(TM) VH | | Descriptor: | Tumor necrosis factor receptor superfamily member 17, UniAb | | Authors: | Clarke, S.C, Ma, B, Trinklein, N.D, Schellenberger, U, Osborn, M, Ouisse, L, Boudreau, A, Davison, L, Harris, K.E, Ugamraj, H, Balasubramani, A, Dang, K, Jorgensen, B, Ogana, H, Pham, D, Pratap, P, Sankaran, P, Anegon, I, van Schooten, W, Bruggemann, M, Buelow, R, Force Aldred, S. | | Deposit date: | 2019-01-18 | | Release date: | 2019-02-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Multispecific Antibody Development Platform Based on Human Heavy Chain Antibodies
Front Immunol, 9, 2018
|
|
7WC0
 
 | |
7LUP
 
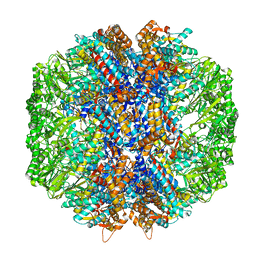 | | Human TRiC/CCT complex with reovirus outer capsid protein sigma-3 | | Descriptor: | Outer capsid protein sigma-3, T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, ... | | Authors: | Knowlton, J.J, Gestaut, D, Ma, B, Taylor, G, Seven, A.B, Leitner, A, Wilson, G.J, Shanker, S, Yates, N.A, Prasad, B.V.V, Aebersold, R, Chiu, W, Frydman, J, Dermody, T.S. | | Deposit date: | 2021-02-22 | | Release date: | 2021-03-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Structural and functional dissection of reovirus capsid folding and assembly by the prefoldin-TRiC/CCT chaperone network.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7WBZ
 
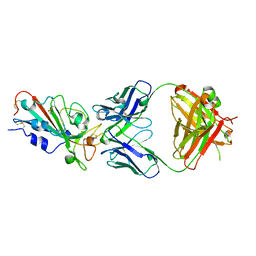 | | Crystal structure of the SARS-Cov-2 RBD in complex with Fab 2303 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2303 heavy chain, 2303 light chain, ... | | Authors: | Xiang, Y, Ma, B. | | Deposit date: | 2021-12-17 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Conformational flexibility in neutralization of SARS-CoV-2 by naturally elicited anti-SARS-CoV-2 antibodies.
Commun Biol, 5, 2022
|
|
7LUM
 
 | | Human TRiC in ATP/AlFx closed state | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Knowlton, J.J, Gestaut, D, Ma, B, Taylor, G, Seven, A.B, Leitner, A, Wilson, G.J, Shanker, S, Yates, N.A, Prasad, B.V.V, Aebersold, R, Chiu, W, Frydman, J, Dermody, T.S. | | Deposit date: | 2021-02-22 | | Release date: | 2021-03-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural and functional dissection of reovirus capsid folding and assembly by the prefoldin-TRiC/CCT chaperone network.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
