6HTZ
 
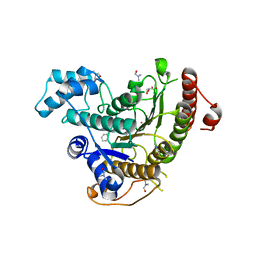 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with a benzohydroxamate inhibitor 8 | | Descriptor: | 4-methoxy-~{N}-oxidanyl-3-(2-phenylethanoylamino)benzamide, DIMETHYLFORMAMIDE, GLYCEROL, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-10-05 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.841 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
6HUB
 
 | | Yeast 20S proteasome with human beta2c (S171G) in complex with 16 | | Descriptor: | (2~{S})-~{N}-[(2~{S},3~{R})-1-[[(2~{S})-1-[4-(aminomethyl)phenyl]-4-methylsulfonyl-butan-2-yl]amino]-3-oxidanyl-1-oxidanylidene-butan-2-yl]-2-[[(2~{R})-2-azido-3-phenyl-propanoyl]amino]-4-methyl-pentanamide, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2018-10-05 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-Based Design of Inhibitors Selective for Human Proteasome beta 2c or beta 2i Subunits.
J.Med.Chem., 62, 2019
|
|
4FMP
 
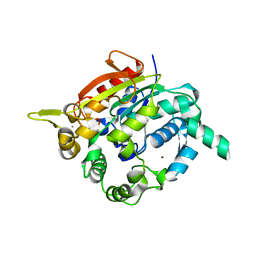 | | Crystal structure of thermostable, organic-solvent tolerant lipase from Geobacillus sp. strain ARM | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Nisbar, N.D, Rahman, R.N.Z.R.A, Ali, M.S.M, Leow, A.T.C. | | Deposit date: | 2012-06-18 | | Release date: | 2013-07-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallization of novel ARM lipase and elucidation of its space-grown crystal structure
Thesis, 2013
|
|
7T80
 
 | |
6HVT
 
 | |
7SZ8
 
 | |
6HWA
 
 | | Yeast 20S proteasome in complex with 43 | | Descriptor: | (2~{S})-~{N}-[(2~{S},3~{R})-1-[(4~{a}~{S},8~{a}~{S})-1,2,3,4,4~{a},5,6,7,8,8~{a}-decahydronaphthalen-2-yl]-4-methyl-3,4-bis(oxidanyl)pentan-2-yl]-3-(4-methoxyphenyl)-2-[[(2~{S})-2-(2-morpholin-4-ylethanoylamino)propanoyl]amino]propanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2018-10-11 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Based Design of Inhibitors Selective for Human Proteasome beta 2c or beta 2i Subunits.
J.Med.Chem., 62, 2019
|
|
6N36
 
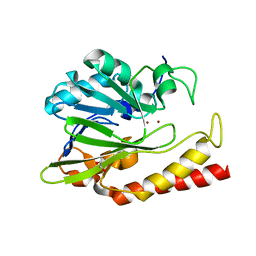 | | Beta-lactamase from Chitinophaga pinensis | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, ZINC ION | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-11-14 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Beta-lactamase from Chitinophaga pinensis
to be published
|
|
7T3S
 
 | |
2MZ7
 
 | |
6HVU
 
 | |
7O6S
 
 | | Crystal structure of a shortened IpgC variant in complex with N-(2H-1,3-benzodioxol-5-ylmethyl)cyclopentanamine | | Descriptor: | CHLORIDE ION, Chaperone protein IpgC, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gardonyi, M, Heine, A, Klebe, G. | | Deposit date: | 2021-04-12 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of a shortened IpgC variant in complex with N-(2H-1,3-benzodioxol-5-ylmethyl)cyclopentanamine
To be published
|
|
7T91
 
 | |
6HWE
 
 | | Yeast 20S proteasome beta2-G45A mutant in complex with carfilzomib | | Descriptor: | (2~{S})-~{N}-[(2~{S})-1-[[(3~{R},4~{S})-2,6-dimethyl-2,3-bis(oxidanyl)heptan-4-yl]amino]-1-oxidanylidene-3-phenyl-propan-2-yl]-4-methyl-2-[[(2~{S})-2-(2-morpholin-4-ylethanoylamino)-4-phenyl-butanoyl]amino]pentanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2018-10-11 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Inhibitors Selective for Human Proteasome beta 2c or beta 2i Subunits.
J.Med.Chem., 62, 2019
|
|
6F70
 
 | | Crystal structure of glutathione transferase Omega 6S from Trametes versicolor | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLUTATHIONE, GLYCEROL, ... | | Authors: | Schwartz, M, Favier, F, Didierjean, C. | | Deposit date: | 2017-12-07 | | Release date: | 2018-06-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Molecular recognition of wood polyphenols by phase II detoxification enzymes of the white rot Trametes versicolor.
Sci Rep, 8, 2018
|
|
1X66
 
 | | Solution structure of the SAM_PNT-domain of the human friend LEUKEMIAINTEGRATION 1 transcription factor | | Descriptor: | Friend leukemia integration 1 transcription factor | | Authors: | Goroncy, A, Kigawa, T, Koshiba, S, Sato, M, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-17 | | Release date: | 2005-11-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SAM_PNT-domain of the human friend LEUKEMIAINTEGRATION 1 transcription factor
To be Published
|
|
6VD1
 
 | | Crystal structure of Arabidopsis thaliana S-adenosylmethionine Synthase 2 (AtMAT2) in complex with S-adenosylmethionine and PPNP | | Descriptor: | (DIPHOSPHONO)AMINOPHOSPHONIC ACID, 1,2-ETHANEDIOL, 1,3-PROPANDIOL, ... | | Authors: | Sekula, B, Ruszkowski, M, Dauter, Z. | | Deposit date: | 2019-12-23 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | S-adenosylmethionine synthases in plants: Structural characterization of type I and II isoenzymes from Arabidopsis thaliana and Medicago truncatula.
Int.J.Biol.Macromol., 151, 2020
|
|
6I0N
 
 | |
6VE3
 
 | | Tetradecameric PilQ from Pseudomonas aeruginosa | | Descriptor: | Fimbrial assembly protein PilQ | | Authors: | McCallum, M, Tammam, S, Rubinstein, J.L, Burrows, L.L, Howell, P.L. | | Deposit date: | 2019-12-28 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | CryoEM map of Pseudomonas aeruginosa PilQ enables structural characterization of TsaP.
Structure, 29, 2021
|
|
6PF9
 
 | | Crystal structure of TS-DHFR from Cryptosporidium hominis in complex with NADPH, FdUMP and 2-(2-(4-((2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl)methyl)benzamido)phenyl)acetic acid. | | Descriptor: | 5-FLUORO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase, METHOTREXATE, ... | | Authors: | Czyzyk, D.J, Valhondo, M, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2019-06-21 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure activity relationship towards design of cryptosporidium specific thymidylate synthase inhibitors.
Eur.J.Med.Chem., 183, 2019
|
|
6VD0
 
 | | Crystal structure of Arabidopsis thaliana S-adenosylmethionine Synthase 2 (AtMAT2) in complex with free Methionine and AMPCPP | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, DI(HYDROXYETHYL)ETHER, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | Sekula, B, Ruszkowski, M, Dauter, Z. | | Deposit date: | 2019-12-23 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | S-adenosylmethionine synthases in plants: Structural characterization of type I and II isoenzymes from Arabidopsis thaliana and Medicago truncatula.
Int.J.Biol.Macromol., 151, 2020
|
|
7TAM
 
 | |
6N91
 
 | | Crystal Structure of Adenosine Deaminase from Vibrio cholerae Complexed with Pentostatin (Deoxycoformycin) | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYCOFORMYCIN, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, ... | | Authors: | Maltseva, N, Kim, Y, Endres, M, Welk, L, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-11-30 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of Adenosine Deaminase from Vibrio cholerae Complexed with Pentostatin (Deoxycoformycin) (CASP target)
To Be Published
|
|
6PAW
 
 | |
6EWK
 
 | | T. californica AChE in complex with a 3-hydroxy-2-pyridine aldoxime. | | Descriptor: | 2-[(~{E})-hydroxyiminomethyl]-6-(5-morpholin-4-ylpentyl)pyridin-3-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | de la Mora, E, Weik, M, Braiki, A, Mougeot, R, Jean, L, Renard, P.I. | | Deposit date: | 2017-11-04 | | Release date: | 2018-11-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Potent 3-Hydroxy-2-Pyridine Aldoxime Reactivators of Organophosphate-Inhibited Cholinesterases with Predicted Blood-Brain Barrier Penetration.
Chemistry, 24, 2018
|
|
