7Q6N
 
 | | Structure of WrbA from Salmonella Typhimurium bound to ME0052 | | Descriptor: | 2-azanyl-4,6-bis(bromanyl)phenol, FLAVIN MONONUCLEOTIDE, NAD(P)H dehydrogenase (quinone), ... | | Authors: | Gabrielsen, M, Beckham, K.S.H, Roe, A.J. | | Deposit date: | 2021-11-08 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structures of WrbA, a spurious target of the salicylidene acylhydrazide inhibitors of type III secretion in Gram-negative pathogens, and verification of improved specificity of next-generation compounds.
Microbiology (Reading, Engl.), 168, 2022
|
|
7Q6M
 
 | | Structure of WrbA from Yersinia pseudotuberculosis in P1 | | Descriptor: | CHLORIDE ION, NAD(P)H dehydrogenase (quinone) | | Authors: | Gabrielsen, M, Beckham, K.S.H, Roe, A.J. | | Deposit date: | 2021-11-08 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structures of WrbA, a spurious target of the salicylidene acylhydrazide inhibitors of type III secretion in Gram-negative pathogens, and verification of improved specificity of next-generation compounds.
Microbiology (Reading, Engl.), 168, 2022
|
|
7QXK
 
 | | As isolated MSOX movie series dataset 1 (0.4 MGy) of the copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, GLYCEROL, ... | | Authors: | Rose, S.L, Baba, S, Okumura, H, Antonyuk, S.V, Sasaki, D, Tosha, T, Kumasaka, T, Eady, R.R, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2022-01-26 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Single crystal spectroscopy and multiple structures from one crystal (MSOX) define catalysis in copper nitrite reductases.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6FAF
 
 | | Crystal structure of human carbonic anhydrase I in complex with the 3-(2,5-dimethylphenyl)-1-(2-hydroxy-5-sulfamoylphenyl)urea inhibitor | | Descriptor: | 1-(2,5-dimethylphenyl)-3-(2-oxidanyl-5-sulfamoyl-phenyl)urea, Carbonic anhydrase 1, GLYCEROL, ... | | Authors: | Ferraroni, M, Supuran, C.T, Bozdag, M, Chiapponi, D. | | Deposit date: | 2017-12-15 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Discovery of 4-Hydroxy-3-(3-(phenylureido)benzenesulfonamides as SLC-0111 Analogues for the Treatment of Hypoxic Tumors Overexpressing Carbonic Anhydrase IX.
J. Med. Chem., 61, 2018
|
|
7QYC
 
 | | As isolated MSOX movie series dataset 20 (8 MGy) of the copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, GLYCEROL, ... | | Authors: | Rose, S.L, Baba, S, Okumura, H, Antonyuk, S.V, Sasaki, D, Tosha, T, Kumasaka, T, Eady, R.R, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2022-01-27 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Single crystal spectroscopy and multiple structures from one crystal (MSOX) define catalysis in copper nitrite reductases.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4ZZY
 
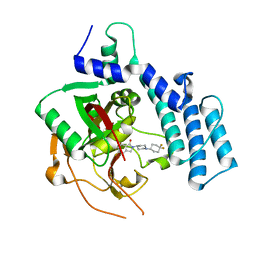 | | Structure of human PARP2 catalytic domain bound to an isoindolinone inhibitor | | Descriptor: | 2-[1-(4,4-Difluorocyclohexyl)-piperidin-4-yl]-6-fluoro-3-oxo-2,3-dihydro-1H-isoindole-4-carboxamide, POLY [ADP-RIBOSE] POLYMERASE 2 | | Authors: | Casale, E, Fasolini, M, Papeo, G, Posteri, H, Borghi, D, Busel, A.A, Caprera, F, Ciomei, M, Cirla, A, Corti, E, DAnello, M, Fasolini, M, Felder, E.R, Forte, B, Galvani, A, Isacchi, A, Khvat, A, Krasavin, M.Y, Lupi, R, Orsini, P, Perego, R, Pesenti, E, Pezzetta, D, Rainoldi, S, RiccardiSirtori, F, Scolaro, A, Sola, F, Zuccotto, F, Donati, D, Montagnoli, A. | | Deposit date: | 2015-04-15 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of 2-[1-(4,4-Difluorocyclohexyl)Piperidin-4-Yl]-6-Fluoro-3-Oxo-2,3-Dihydro-1H-Isoindole-4-Carboxamide (Nms-P118): A Potent, Orally Available and Highly Selective Parp- 1 Inhibitor for Cancer Therapy.
J.Med.Chem., 58, 2015
|
|
7QC6
 
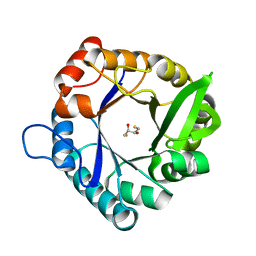 | | HisF_C9A_L50H_I52H mutant (apo) from T. maritima | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Imidazole glycerol phosphate synthase subunit HisF | | Authors: | Beaumet, M, Dose, A, Braeuer, A, Mahy, J, Ghattas, W, Groll, M, Hess, C. | | Deposit date: | 2021-11-22 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An artificial metalloprotein with metal-adaptive coordination sites and Ni-dependent quercetinase activity.
J.Inorg.Biochem., 235, 2022
|
|
7QC7
 
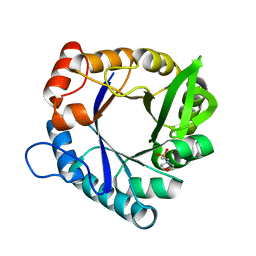 | | HisF-C9A-D11E-V33A_L50H_I52H mutant (apo) from T. maritima | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Imidazole glycerol phosphate synthase subunit HisF | | Authors: | Beaumet, M, Dose, A, Braeuer, A, Mahy, J, Ghattas, W, Groll, M, Hess, C. | | Deposit date: | 2021-11-22 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An artificial metalloprotein with metal-adaptive coordination sites and Ni-dependent quercetinase activity.
J.Inorg.Biochem., 235, 2022
|
|
7QC8
 
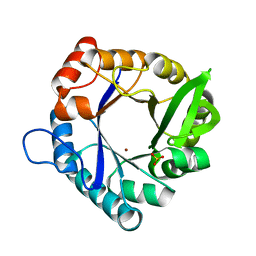 | | HisF-C9A-D11E-V33A_L50H_I52H mutant in complex with Zn(II) from T. maritima | | Descriptor: | Imidazole glycerol phosphate synthase subunit HisF, SULFATE ION, ZINC ION | | Authors: | Beaumet, M, Dose, A, Braeuer, A, Mahy, J, Ghattas, W, Groll, M, Hess, C. | | Deposit date: | 2021-11-22 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An artificial metalloprotein with metal-adaptive coordination sites and Ni-dependent quercetinase activity.
J.Inorg.Biochem., 235, 2022
|
|
7QC9
 
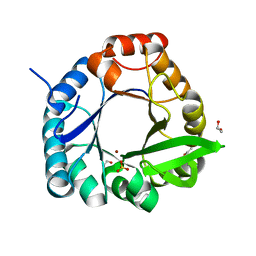 | | HisF-C9A-D11E-V33A_L50H_I52H mutant in complex with Ni(II) from T. maritima | | Descriptor: | 1,2-ETHANEDIOL, Imidazole glycerol phosphate synthase subunit HisF, NICKEL (II) ION, ... | | Authors: | Beaumet, M, Dose, A, Braeuer, A, Mahy, J, Ghattas, W, Groll, M, Hess, C. | | Deposit date: | 2021-11-22 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An artificial metalloprotein with metal-adaptive coordination sites and Ni-dependent quercetinase activity.
J.Inorg.Biochem., 235, 2022
|
|
6JLK
 
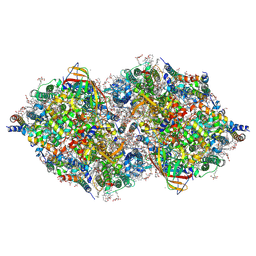 | | XFEL structure of cyanobacterial photosystem II (1F state, dataset1) | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Shen, J.R. | | Deposit date: | 2019-03-06 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | An oxyl/oxo mechanism for oxygen-oxygen coupling in PSII revealed by an x-ray free-electron laser.
Science, 366, 2019
|
|
6JLM
 
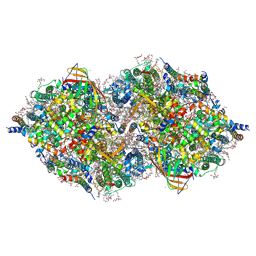 | | XFEL structure of cyanobacterial photosystem II (dark state, dataset2) | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Shen, J.R. | | Deposit date: | 2019-03-06 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | An oxyl/oxo mechanism for oxygen-oxygen coupling in PSII revealed by an x-ray free-electron laser.
Science, 366, 2019
|
|
7ZQK
 
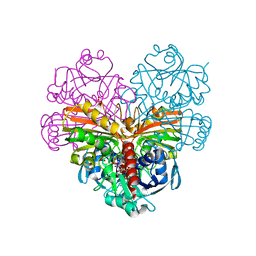 | | Crystal structure of photosynthetic glyceraldehyde-3-phosphate dehydrogenase from Chlamydomonas reinhardtii (CrGAPA) complexed with NAD+ | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Fermani, S, Zaffagnini, M, Lemaire, S.D, Falini, G, Fanti, S, Rossi, J. | | Deposit date: | 2022-04-30 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural snapshots of nitrosoglutathione binding and reactivity underlying S-nitrosylation of photosynthetic GAPDH.
Redox Biol, 54, 2022
|
|
7ZQ4
 
 | | Crystal structure of photosynthetic glyceraldehyde-3-phosphate dehydrogenase from Chlamydomonas reinhardtii (CrGAPA) complexed with NADP+ and the oxidated catalytic cysteine | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Glyceraldehyde-3-phosphate dehydrogenase A, ... | | Authors: | Fermani, S, Zaffagnini, M, Lemaire, S.D, Falini, G, Fanti, S, Rossi, J. | | Deposit date: | 2022-04-29 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural snapshots of nitrosoglutathione binding and reactivity underlying S-nitrosylation of photosynthetic GAPDH.
Redox Biol, 54, 2022
|
|
5FUI
 
 | | Crystal structure of the C-terminal CBM6 of LamC a marine laminarianse from Zobellia galactanivorans | | Descriptor: | 2-AMINOMETHYL-PYRIDINE, ENDO-1,3-BETA-GLUCANASE, FAMILY GH16, ... | | Authors: | Labourel, A, Jam, M, Legentil, L, Sylla, B, Hehemann, J.H, Ficko-Blean, E, Ferrieres, V, Czjzek, M, Michel, G. | | Deposit date: | 2016-01-27 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Unraveling the Multivalent Binding of a Marine Family 6 Carbohydrate-Binding Module with its Native Laminarin Ligand.
FEBS J., 283, 2016
|
|
6BAD
 
 | |
7Q94
 
 | | Crystal Structure of Agrobacterium tumefaciens NADQ, DNA complex. | | Descriptor: | DNA binding region (31-MER), NADQ transcription factor | | Authors: | Cianci, M, Minazzato, G, Heroux, A, Raffaelli, N, Sorci, L, Gasparrini, M. | | Deposit date: | 2021-11-11 | | Release date: | 2022-11-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Bacterial NadQ (COG4111) is a Nudix-like, ATP-responsive regulator of NAD biosynthesis.
J.Struct.Biol., 214, 2022
|
|
7QC3
 
 | | HisF from T. maritima | | Descriptor: | Imidazole glycerol phosphate synthase subunit HisF | | Authors: | Beaumet, M, Dose, A, Braeuer, A, Mahy, J, Ghattas, W, Groll, M, Hess, C. | | Deposit date: | 2021-11-22 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | An artificial metalloprotein with metal-adaptive coordination sites and Ni-dependent quercetinase activity.
J.Inorg.Biochem., 235, 2022
|
|
6JPP
 
 | |
8J1K
 
 | | co-crystal structure of non-carboxylic acid inhibitor with PHD2 | | Descriptor: | Egl nine homolog 1, MANGANESE (II) ION, N-[(6-cyanopyridin-3-yl)methyl]-5-oxidanyl-2-[(3R)-3-oxidanylpyrrolidin-1-yl]-1,7-naphthyridine-6-carboxamide | | Authors: | Xu, J, Fu, Y, Ding, X, Meng, Q, Wang, L, Zhang, M, Ding, X, Ren, F, Zhavoronkov, A. | | Deposit date: | 2023-04-13 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.451 Å) | | Cite: | co-crystal structure of non-carboxylic acid inhibitor with PHD2
To Be Published
|
|
5A50
 
 | |
7Q91
 
 | | Crystal Structure of Agrobacterium tumefaciens NADQ, native form. | | Descriptor: | NADQ transcription factor, SODIUM ION | | Authors: | Cianci, M, Minazzato, G, Heroux, A, Raffaelli, N, Sorci, L, Gasparrini, M. | | Deposit date: | 2021-11-11 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Bacterial NadQ (COG4111) is a Nudix-like, ATP-responsive regulator of NAD biosynthesis.
J.Struct.Biol., 214, 2022
|
|
5AGT
 
 | | Crystal structure of the LeuRS editing domain of Mycobacterium tuberculosis in complex with the adduct (S)-3-(Aminomethyl)-4-chloro-7-ethoxybenzo[c][1,2]oxaborol-1(3H)-ol-AMP | | Descriptor: | 4-Chloro-3-aminomethyl-7-[ethoxy]-3H-benzo[C][1,2]oxaborol-1-ol modified adenosine, GLYCEROL, LEUCINE--TRNA LIGASE, ... | | Authors: | Palencia, A, Li, X, Alley, M.R.K, Ding, C, Easom, E.E, Hernandez, V, Meewan, M, Mohan, M, Rock, F.L, Franzblau, S.G, Wang, Y, Lenaerts, A.J, Parish, T, Cooper, C.B, Waters, M.G, Ma, Z, Mendoza, A, Barros, D, Cusack, S, Plattner, J.J. | | Deposit date: | 2015-02-03 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Discovery of Novel Oral Protein Synthesis Inhibitors of Mycobacterium Tuberculosis that Target Leucyl-tRNA Synthetase.
Antimicrob.Agents Chemother., 60, 2016
|
|
7F8T
 
 | | Re-refinement of the 2XRY X-ray structure of archaeal class II CPD photolyase from Methanosarcina mazei | | Descriptor: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.C, Nango, E, Gusti-Ngurah-Putu, E.-P, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Sugahara, M, Owada, S, Joti, Y, Tanaka, R, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-07-02 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
5FOT
 
 | | HUMANISED MONOMERIC RADA IN COMPLEX WITH FHTU TETRAPEPTIDE | | Descriptor: | DNA REPAIR AND RECOMBINATION PROTEIN RADA, FHTU PEPTIDE, PHOSPHATE ION | | Authors: | Scott, D.E, Marsh, M, Blundell, T.L, Abell, C, Hyvonen, M. | | Deposit date: | 2015-11-26 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.189 Å) | | Cite: | Structure Activity Relationship of the Peptide Binding Motif Mediating the Rad51:Brca2 Protein-Protein Interaction.
FEBS Lett., 590, 2016
|
|
