8RR2
 
 | | 3-keto-glycoside eliminase/hydratase in komplex with alpha-3-keto-glucose | | Descriptor: | (2~{R},3~{R},5~{S},6~{S})-2-(hydroxymethyl)-3,5,6-tris(oxidanyl)oxan-4-one, 1,2-ETHANEDIOL, 3-keto-disaccharide hydrolase domain-containing protein, ... | | Authors: | Pfeiffer, M, Kastner, K, Oberdorfer, G, Nidetzky, B. | | Deposit date: | 2024-01-22 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Enzyme Machinery for Bacterial Glucoside Metabolism through a Conserved Non-hydrolytic Pathway.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
6KNP
 
 | |
8RKD
 
 | | TadA/CpaF with AMPPNP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Hohl, M, Low, H. | | Deposit date: | 2023-12-24 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Bidirectional pilus processing in the Tad pilus system motor CpaF.
Nat Commun, 15, 2024
|
|
7TDE
 
 | | AtTPC1 DDE mutant with 1 mM Ca2+ | | Descriptor: | Two pore calcium channel protein 1 | | Authors: | Dickinson, M.S, Stroud, R.M. | | Deposit date: | 2021-12-31 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular basis of multistep voltage activation in plant two-pore channel 1.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6KN3
 
 | |
4QOI
 
 | | Crystal structure of FMN quinone reductase 2 in complex with melatonin at 1.55A | | Descriptor: | FLAVIN MONONUCLEOTIDE, N-[2-(5-methoxy-1H-indol-3-yl)ethyl]acetamide, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Serriere, J, Boutin, J.A, Isabet, T, Antoine, M, Ferry, G. | | Deposit date: | 2014-06-20 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of FMN quinone reductase 2 in complex with melatonin at 1.55A
To be Published
|
|
8RZK
 
 | | The Michaelis complex of ZgGH129 D486N from Zobellia galactanivorans with neo-b/k-oligo-carrageenan tetrasaccharide (beta-kappa neo-oligo-carrageenan DP4). | | Descriptor: | 1,2-ETHANEDIOL, 3,6-anhydro-alpha-D-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-D-galactopyranose-(1-3)-4-O-sulfo-beta-D-galactopyranose, CHLORIDE ION, ... | | Authors: | Roret, T, Czjzek, M, Ficko-Blean, E. | | Deposit date: | 2024-02-12 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Constrained Catalytic Itinerary of a Retaining 3,6-Anhydro-D-Galactosidase, a Key Enzyme in Red Algal Cell Wall Degradation.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
6KN2
 
 | |
8RO4
 
 | | The crystal structure of 2-hydroxy-3-keto-glucal hydratase AtHYD from A. tumefaciens | | Descriptor: | 2-hydroxy-3-keto-glucal hydratase, MANGANESE (II) ION | | Authors: | Grininger, C, Bitter, J, Pfeiffer, M, Nidetzky, B, Pavkov-Keller, T. | | Deposit date: | 2024-01-11 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Enzyme Machinery for Bacterial Glucoside Metabolism through a Conserved Non-hydrolytic Pathway.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
3OKU
 
 | | Human Carbonic Anhydrase II in complex with 2-Ethylestrone-3-O-sulfamate | | Descriptor: | (9beta)-2-ethyl-17-oxoestra-1(10),2,4-trien-3-yl sulfamate, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Sippel, K.H, Stander, B.A, Robbins, A.H, Tu, C.K, Agbandje-McKenna, M, Silverman, D.N, Joubert, A.M, McKenna, R. | | Deposit date: | 2010-08-25 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Characterization of Carbonic Anhydrase Isozyme Specific Inhibition by Sulfamated 2-Ethylestra Compounds
LETT.DRUG DES.DISCOVERY, 8, 2011
|
|
4QPB
 
 | | Catalytic domain of the antimicrobial peptidase lysostaphin from Staphylococcus simulans crystallized in the absence of phosphate | | Descriptor: | 1,2-ETHANEDIOL, Lysostaphin, ZINC ION | | Authors: | Sabala, I, Jagielska, E, Bardelang, P.T, Czapinska, H, Dahms, S.O, Sharpe, J.A, James, R, Than, M.E, Thomas, N.R, Bochtler, M. | | Deposit date: | 2014-06-22 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of the antimicrobial peptidase lysostaphin from Staphylococcus simulans.
Febs J., 281, 2014
|
|
4QI0
 
 | | X-ray structure of the ROQ domain from murine Roquin-1 | | Descriptor: | 1,2-ETHANEDIOL, Roquin-1 | | Authors: | Janowski, R, Schlundt, A, Sattler, M, Niessing, D. | | Deposit date: | 2014-05-30 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural basis for RNA recognition in roquin-mediated post-transcriptional gene regulation.
Nat.Struct.Mol.Biol., 21, 2014
|
|
8RJF
 
 | | TadA/CpaF with ADP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Hohl, M, Low, H. | | Deposit date: | 2023-12-21 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Bidirectional pilus processing in the Tad pilus system motor CpaF.
Nat Commun, 15, 2024
|
|
3OIM
 
 | | Human Carbonic anhydrase II bound by 2-Ethylestradiol 3-O-sulfamate | | Descriptor: | (14beta,17alpha)-2-ethyl-17-hydroxyestra-1(10),2,4-trien-3-yl sulfamate, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Sippel, K.H, Stander, B.A, Robbins, A.H, Tu, C.K, Agbandje-McKenna, M, Silverman, D.N, Joubert, A.M, McKenna, R. | | Deposit date: | 2010-08-19 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Characterization of Carbonic Anhydrase Isozyme Specific Inhibition by Sulfamated 2-Ethylestra Compounds
LETT.DRUG DES.DISCOVERY, 8, 2011
|
|
5MC4
 
 | | Crystal Structure of Gly448Arg mutant of Human Prolidase with Mn ions and GlyPro ligand | | Descriptor: | GLYCEROL, GLYCINE, HYDROXIDE ION, ... | | Authors: | Wilk, P, Mueller, U, Dobbek, H, Weiss, M.S. | | Deposit date: | 2016-11-09 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for prolidase deficiency disease mechanisms.
FEBS J., 285, 2018
|
|
4QOG
 
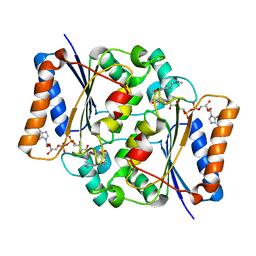 | | Crystal structure of fad quinone reductase 2 in complex with melatonin at 1.4A | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, N-[2-(5-methoxy-1H-indol-3-yl)ethyl]acetamide, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Serriere, J, Boutin, J.A, Isabet, T, Antoine, M, Ferry, G. | | Deposit date: | 2014-06-20 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of fad quinone reductase 2 in complex with melatonin at 1.4A
To be Published
|
|
8RKL
 
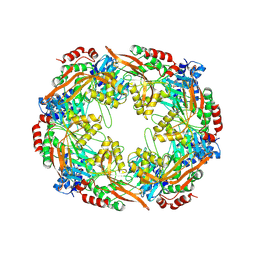 | | TadA/CpaF nucleotide free | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Pilus assembly ATPase CpaF | | Authors: | Hohl, M, Low, H. | | Deposit date: | 2023-12-26 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Bidirectional pilus processing in the Tad pilus system motor CpaF.
Nat Commun, 15, 2024
|
|
4QNS
 
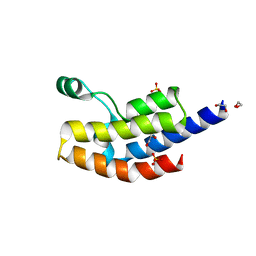 | | Crystal structure of bromodomain from Plasmodium faciparum GCN5, PF3D7_0823300 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Histone acetyltransferase GCN5, ... | | Authors: | Fonseca, M, Tallant, C, Knapp, S, Loppnau, P, von Delft, F, Wernimont, A.K, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-18 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.497 Å) | | Cite: | Crystal structure of bromodomain from Plasmodium faciparum GCN5, PF3D7_0823300
TO BE PUBLISHED
|
|
8RFB
 
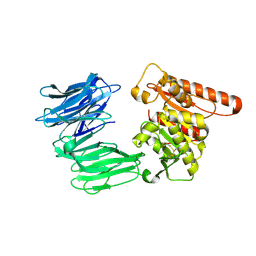 | | Cryo-EM structure of the R243C mutant of human Prolyl Endopeptidase-Like (PREPL) protein involved in Congenital myasthenic syndrome-22 (CMS22) | | Descriptor: | Prolyl endopeptidase-like | | Authors: | Theodoropoulou, A, Cavani, E, Antanasijevic, A, Marcaida, M.J, Dal Peraro, M. | | Deposit date: | 2023-12-12 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.01 Å) | | Cite: | Missense variants in CMS22 patients reveal that PREPL has both enzymatic and nonenzymatic functions.
JCI Insight, 9, 2024
|
|
8RFZ
 
 | |
4QOF
 
 | | Crystal structure of fmn quinone reductase 2 AT 1.55A | | Descriptor: | FLAVIN MONONUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ZINC ION | | Authors: | Serriere, J, Boutin, J.A, Isabet, T, Antoine, M, Ferry, G. | | Deposit date: | 2014-06-20 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of FMN quinone reductase 2 at 1.55A
To be Published
|
|
8RII
 
 | |
8RK0
 
 | | HCV E1/E2 homodimer complex, ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HCV E1, ... | | Authors: | Augestad, E.H, Olesen, C.H, Groenberg, C, Soerensen, A, Velazquez-Moctezuma, R, Fanalista, M, Bukh, J, Wang, K, Gourdon, P, Prentoe, J. | | Deposit date: | 2023-12-22 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | The hepatitis C virus envelope protein complex is a dimer of heterodimers.
Nature, 633, 2024
|
|
5MSC
 
 | |
5MSQ
 
 | |
