7EGO
 
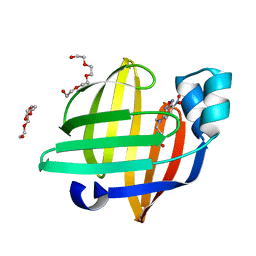 | | X-ray structure of the human heart fatty acid-binding protein complexed with the fluorescent probe HA527 | | Descriptor: | 3-[methyl-(4-nitro-2,1,3-benzoxadiazol-7-yl)amino]propanoic acid, Fatty acid-binding protein, heart, ... | | Authors: | Takabayashi, M, Yokota, J, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Sugiyama, S. | | Deposit date: | 2021-03-24 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | X-ray structure of the human heart fatty acid-binding protein complexed with the fluorescent probe HA527
To Be Published
|
|
3E9U
 
 | | Crystal structure of Calx CBD2 domain | | Descriptor: | Na/Ca exchange protein | | Authors: | Wu, M, Zheng, L. | | Deposit date: | 2008-08-23 | | Release date: | 2009-01-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of CBD2 from the Drosophila Na(+)/Ca(2+) exchanger: diversity of Ca(2+) regulation and its alternative splicing modification.
J.Mol.Biol., 387, 2009
|
|
8Y6F
 
 | |
3RBS
 
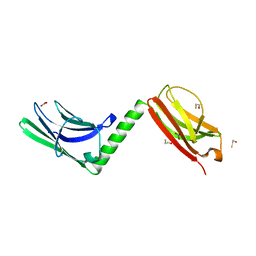 | |
3RIE
 
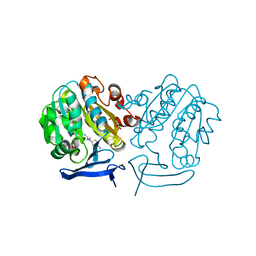 | | The structure of Plasmodium falciparum spermidine synthase in complex with 5'-methylthioadenosine and N-(3-aminopropyl)-trans-cyclohexane-1,4-diamine | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 5'-DEOXY-5'-METHYLTHIOADENOSINE, GLYCEROL, ... | | Authors: | Williams, M, Sprenger, J, Persson, L, Louw, A.I, Birkholtz, L, Al-Karadaghi, S. | | Deposit date: | 2011-04-13 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A novel inhibitor of Plasmodium falciparum spermidine synthase: a twist in the tail.
Malar J, 14, 2015
|
|
8YRJ
 
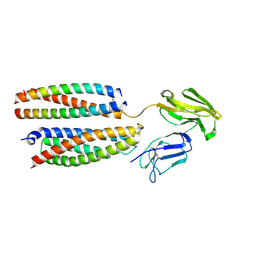 | | Mouse Fc epsilon RI | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, High affinity immunoglobulin epsilon receptor subunit alpha, High affinity immunoglobulin epsilon receptor subunit beta, ... | | Authors: | Zhang, Z, Yui, M, Ohto, U, Shimizu, T. | | Deposit date: | 2024-03-21 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Mouse Fc epsilon RI
To Be Published
|
|
6JZU
 
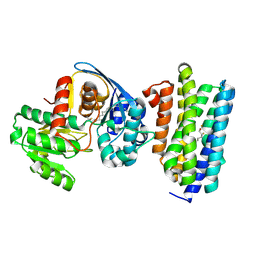 | | The crystal structure of acyl-acyl carrier protein (acyl-ACP) reductase (AAR) in complex with aldehyde deformylating oxygenase (ADO) | | Descriptor: | Aldehyde decarbonylase, FE (II) ION, HEXADECAN-1-OL, ... | | Authors: | Zhang, H.M, Li, M, Gao, Y. | | Deposit date: | 2019-05-03 | | Release date: | 2020-04-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.181 Å) | | Cite: | Structural insights into catalytic mechanism and product delivery of cyanobacterial acyl-acyl carrier protein reductase.
Nat Commun, 11, 2020
|
|
8XLP
 
 | | Structure of inactive Photosystem II associated with CAC antenna from Rhodomonas Salina | | Descriptor: | (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E})-3,7,12,16-tetramethyl-18-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-3,5,7,9,11,13,15-heptaen-1,17-diynyl]cyclohex-3-en-1-ol, (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-(2,6,6-trimethylcyclohexen-1-yl)octadeca-3,5,7,9,11,13,15,17-octaen-1-ynyl]cyclohex-3-en-1-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Si, L, Li, M. | | Deposit date: | 2023-12-26 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Structural basis for the distinct core-antenna assembly of cryptophyte photosystem II.
Nat Commun, 15, 2024
|
|
3B42
 
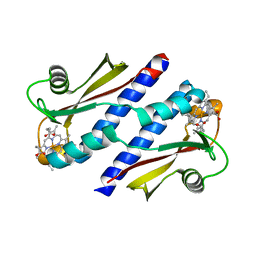 | | Periplasmic sensor domain of chemotaxis protein GSU0935 | | Descriptor: | Methyl-accepting chemotaxis protein, putative, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Pokkuluri, P.R, Schiffer, M. | | Deposit date: | 2007-10-23 | | Release date: | 2008-04-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures and solution properties of two novel periplasmic sensor domains with c-type heme from chemotaxis proteins of Geobacter sulfurreducens: implications for signal transduction.
J.Mol.Biol., 377, 2008
|
|
3B2Z
 
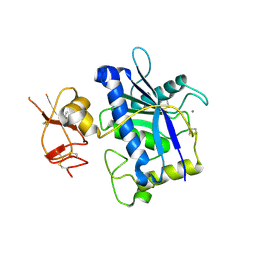 | | Crystal Structure of ADAMTS4 (apo form) | | Descriptor: | ADAMTS-4, CALCIUM ION, ZINC ION | | Authors: | Mosyak, L, Stahl, M, Somers, W. | | Deposit date: | 2007-10-19 | | Release date: | 2007-12-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of the two major aggrecan degrading enzymes, ADAMTS4 and ADAMTS5.
Protein Sci., 17, 2008
|
|
3B8P
 
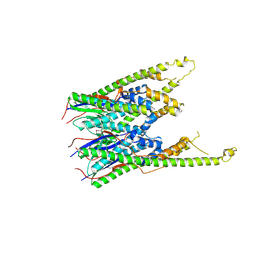 | |
3B9M
 
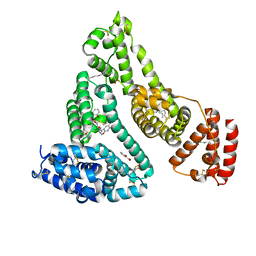 | | Human serum albumin complexed with myristate, 3'-azido-3'-deoxythymidine (AZT) and salicylic acid | | Descriptor: | 2-HYDROXYBENZOIC ACID, 3'-azido-3'-deoxythymidine, MYRISTIC ACID, ... | | Authors: | Zhu, L, Yang, F, Chen, L, Meehan, E.J, Huang, M. | | Deposit date: | 2007-11-05 | | Release date: | 2008-05-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A new drug binding subsite on human serum albumin and drug-drug interaction studied by X-ray crystallography
J.Struct.Biol., 162, 2008
|
|
3BAZ
 
 | | Structure of hydroxyphenylpyruvate reductase from coleus blumei in complex with NADP+ | | Descriptor: | Hydroxyphenylpyruvate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Janiak, V, Klebe, G, Petersen, M, Heine, A. | | Deposit date: | 2007-11-09 | | Release date: | 2008-11-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and substrate docking of a hydroxy(phenyl)pyruvate reductase from the higher plant Coleus blumei Benth
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3RK4
 
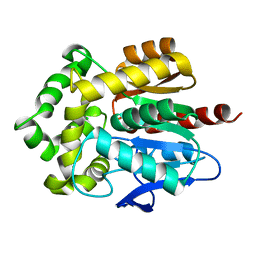 | | Structure of Rhodococcus rhodochrous haloalkane dehalogenase mutant DhaA31 | | Descriptor: | CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Lahoda, M, Stsiapanava, A, Mesters, J, Chaloupkova, R, Damborsky, J, Kuta Smatanova, I. | | Deposit date: | 2011-04-17 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Crystallographic analysis of 1,2,3-trichloropropane biodegradation by the haloalkane dehalogenase DhaA31.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6KIP
 
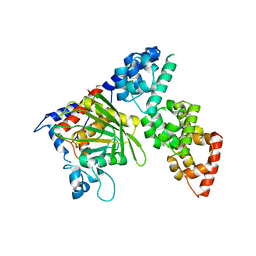 | |
3BA1
 
 | | Structure of hydroxyphenylpyruvate reductase from coleus blumei | | Descriptor: | Hydroxyphenylpyruvate reductase | | Authors: | Janiak, V, Klebe, G, Petersen, M, Heine, A. | | Deposit date: | 2007-11-07 | | Release date: | 2008-11-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structure and substrate docking of a hydroxy(phenyl)pyruvate reductase from the higher plant Coleus blumei Benth.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
6IQY
 
 | | High resolution structure of bilirubin oxidase from Myrothecium verrucaria - M467Q mutant, anaerobically prepared | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Bilirubin oxidase, ... | | Authors: | Shibata, N, Akter, M, Higuchi, Y. | | Deposit date: | 2018-11-09 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Redox Potential-Dependent Formation of an Unusual His-Trp Bond in Bilirubin Oxidase.
Chemistry, 24, 2018
|
|
6IO4
 
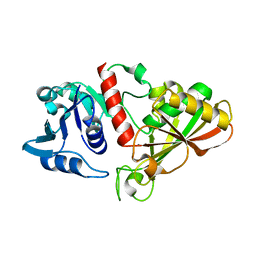 | |
6IOJ
 
 | |
6I47
 
 | | Structure of P. aeruginosa LpxC with compound 10: (2RS)-4-(5-(2-Fluoro-4-methoxyphenyl)-1-oxoisoindolin-2-yl)-N-hydroxy-2-methyl-2-(methylsulfonyl)butanamide | | Descriptor: | (2~{R})-4-[6-(2-fluoranyl-4-methoxy-phenyl)-3-oxidanylidene-1~{H}-isoindol-2-yl]-2-methyl-2-methylsulfonyl-~{N}-oxidanyl-butanamide, (2~{S})-4-[6-(2-fluoranyl-4-methoxy-phenyl)-3-oxidanylidene-1~{H}-isoindol-2-yl]-2-methyl-2-methylsulfonyl-~{N}-oxidanyl-butanamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ... | | Authors: | Surivet, J.-P, Panchaud, P, Specklin, J.-L, Diethelm, S, Blumstein, A.-C, Gauvin, J.-C, Jacob, L, Masse, F, Mathieu, G, Mirre, A, Schmitt, C, Enderlin-Paput, M, Lange, R, Bur, D, Tidten-Luksch, N, Gnerre, C, Seeland, S, Hermann, C, Locher, H.H, Seiler, P, Mac Sweeney, A, Hubschwerlen, C, Ritz, D, Rueedi, G. | | Deposit date: | 2018-11-09 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Novel Inhibitors of LpxC Displaying Potent in Vitro Activity against Gram-Negative Bacteria.
J.Med.Chem., 63, 2020
|
|
6K0A
 
 | | cryo-EM structure of an archaeal Ribonuclease P | | Descriptor: | 50S ribosomal protein L7Ae, RPR, Ribonuclease P protein component 1, ... | | Authors: | Wan, F, Lan, P, Wu, J, Lei, M. | | Deposit date: | 2019-05-05 | | Release date: | 2019-06-19 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Cryo-electron microscopy structure of an archaeal ribonuclease P holoenzyme.
Nat Commun, 10, 2019
|
|
3O73
 
 | | Crystal structure of quinone reductase 2 in complex with the indolequinone MAC627 | | Descriptor: | 5-[(4-aminobutyl)amino]-1,2-dimethyl-3-[(4-nitrophenoxy)methyl]-1H-indole-4,7-dione, FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Dufour, M, Yan, C, Colucci, M.A, Siegel, D, Li, Y, De Matteis, C.I, Ross, D, Moody, C.J. | | Deposit date: | 2010-07-30 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism-Based Inhibition of Quinone Reductase 2 (NQO2): Selectivity for NQO2 over NQO1 and Structural Basis for Flavoprotein Inhibition.
Chembiochem, 12, 2011
|
|
3O1C
 
 | | High resolution crystal structure of histidine triad nucleotide-binding protein 1 (Hint1) C38A mutant from rabbit complexed with Adenosine | | Descriptor: | ADENOSINE, Histidine triad nucleotide-binding protein 1, SODIUM ION | | Authors: | Dolot, R.M, Ozga, M, Krakowiak, A.K, Nawrot, B, Stec, W.J. | | Deposit date: | 2010-07-21 | | Release date: | 2010-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Histidine Triad Nucleotide-binding Protein 1 (HINT-1) Phosphoramidase Transforms Nucleoside 5'-O-Phosphorothioates to Nucleoside 5'-O-Phosphates.
J.Biol.Chem., 285, 2010
|
|
3O1X
 
 | | High resolution crystal structure of histidine triad nucleotide-binding protein 1 (Hint1) C84A mutant from rabbit complexed with adenosine | | Descriptor: | ADENOSINE, Histidine triad nucleotide-binding protein 1, SODIUM ION | | Authors: | Dolot, R.M, Ozga, M, Krakowiak, A.K, Nawrot, B, Stec, W.J. | | Deposit date: | 2010-07-22 | | Release date: | 2010-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Histidine Triad Nucleotide-binding Protein 1 (HINT-1) Phosphoramidase Transforms Nucleoside 5'-O-Phosphorothioates to Nucleoside 5'-O-Phosphates.
J.Biol.Chem., 285, 2010
|
|
3O1Z
 
 | | High resolution crystal structure of histidine triad nucleotide-binding protein 1 (Hint1) double cysteine mutant from rabbit | | Descriptor: | Histidine triad nucleotide-binding protein 1 | | Authors: | Dolot, R.M, Ozga, M, Krakowiak, A.K, Nawrot, B, Stec, W.J. | | Deposit date: | 2010-07-22 | | Release date: | 2010-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Histidine Triad Nucleotide-binding Protein 1 (HINT-1) Phosphoramidase Transforms Nucleoside 5'-O-Phosphorothioates to Nucleoside 5'-O-Phosphates.
J.Biol.Chem., 285, 2010
|
|
