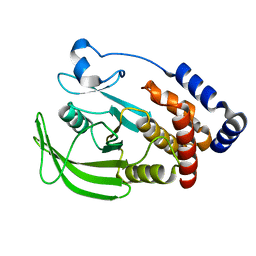
5KA4
 
 | |
5KAB
 
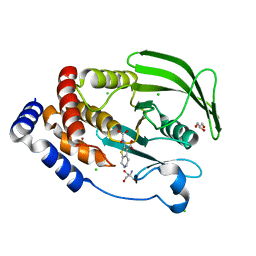 | | Protein Tyrosine Phosphatase 1B Delta helix 7, P185G mutant in complex with TCS401, open state | | Descriptor: | 2-(OXALYL-AMINO)-4,5,6,7-TETRAHYDRO-THIENO[2,3-C]PYRIDINE-3-CARBOXYLIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Choy, M.S, Machado, L.E.S.F, Peti, W, Page, R. | | Deposit date: | 2016-06-01 | | Release date: | 2017-03-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.968 Å) | | Cite: | Conformational Rigidity and Protein Dynamics at Distinct Timescales Regulate PTP1B Activity and Allostery.
Mol. Cell, 65, 2017
|
|
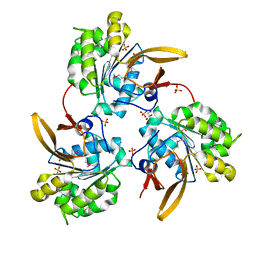
4WIA
 
 | |
8H3M
 
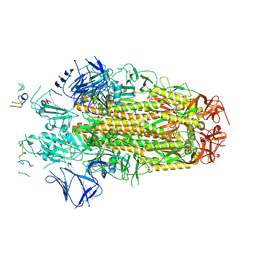 | | Conformation 1 of SARS-CoV-2 Omicron BA.1 Variant Spike protein complexed with MO1 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MO1 heavy chain, Spike glycoprotein | | Authors: | Ishimaru, H, Nishimura, M, Sutandhio, S, Shigematsu, H, Kato, K, Hasegawa, N, Mori, Y. | | Deposit date: | 2022-10-09 | | Release date: | 2023-05-10 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Identification and Analysis of Monoclonal Antibodies with Neutralizing Activity against Diverse SARS-CoV-2 Variants.
J.Virol., 97, 2023
|
|
4WLS
 
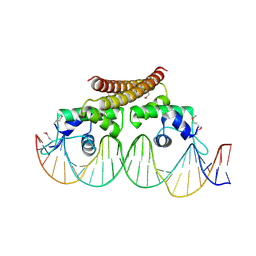 | | Crystal structure of the metal-free (repressor) form of E. Coli CUER, a copper efflux regulator, bound to COPA promoter DNA | | Descriptor: | COPA PROMOTER DNA NON-TEMPLATE STRAND, COPA PROMOTER DNA NON-TEMPLATE STRAND (ALTERNATE CONFORMATION), COPA PROMOTER DNA TEMPLATE STRAND, ... | | Authors: | Philips, S.J, Canalizo-Hernandez, M, Mondragon, A, O'Halloran, T.V. | | Deposit date: | 2014-10-08 | | Release date: | 2015-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | Allosteric transcriptional regulation via changes in the overall topology of the core promoter.
Science, 349, 2015
|
|
8H3N
 
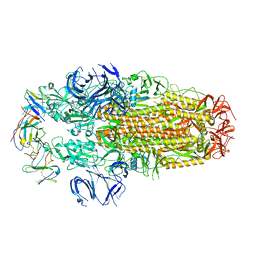 | | Conformation 2 of SARS-CoV-2 Omicron BA.1 Variant Spike protein complexed with MO1 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MO1 heavy-chain, MO1 light chain, ... | | Authors: | Ishimaru, H, Nishimura, M, Sutandhio, S, Shigematsu, H, Kato, K, Hasegawa, N, Mori, Y. | | Deposit date: | 2022-10-09 | | Release date: | 2023-05-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Identification and Analysis of Monoclonal Antibodies with Neutralizing Activity against Diverse SARS-CoV-2 Variants.
J.Virol., 97, 2023
|
|
4X3B
 
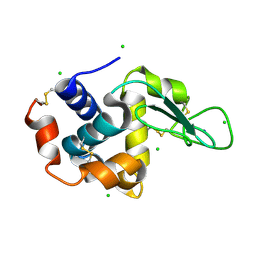 | | A micro-patterned silicon chip as sample holder for macromolecular crystallography experiments with minimal background scattering | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Roedig, P, Vartiainen, I, Duman, R, Panneerselvam, S, Stuebe, N, Lorbeer, O, Warmer, M, Sutton, G, Stuart, D.I, Weckert, E, David, C, Wagner, A, Meents, A. | | Deposit date: | 2014-11-28 | | Release date: | 2015-06-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A micro-patterned silicon chip as sample holder for macromolecular crystallography experiments with minimal background scattering.
Sci Rep, 5, 2015
|
|
5Y80
 
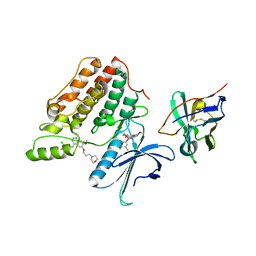 | | Complex structure of cyclin G-associated kinase with gefitinib | | Descriptor: | Cyclin-G-associated kinase, Gefitinib, NANOBODY | | Authors: | Ohbayashi, N, Murayama, K, Kato-Murayama, M, Shirouzu, M. | | Deposit date: | 2017-08-18 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for the Inhibition of Cyclin G-Associated Kinase by Gefitinib.
ChemistryOpen, 7, 2018
|
|
4X9E
 
 | | DEOXYGUANOSINETRIPHOSPHATE TRIPHOSPHOHYDROLASE from Escherichia coli with two DNA effector molecules | | Descriptor: | Deoxyguanosinetriphosphate triphosphohydrolase, MAGNESIUM ION, RNA (5'-R(P*CP*CP*C)-3') | | Authors: | Singh, D, Gawel, D, Itsko, M, Krahn, J.M, London, R.E, Schaaper, R.M. | | Deposit date: | 2014-12-11 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of Escherichia coli dGTP Triphosphohydrolase: A HEXAMERIC ENZYME WITH DNA EFFECTOR MOLECULES.
J.Biol.Chem., 290, 2015
|
|
4WEM
 
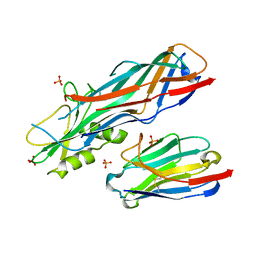 | | Co-complex structure of the F4 fimbrial adhesin FaeG variant ac with llama single domain antibody V1 | | Descriptor: | Anti-F4+ETEC bacteria VHH variable region, K88 fimbrial protein AC, PHOSPHATE ION | | Authors: | Moonens, K, Van den Broeck, I, Pardon, E, De Kerpel, M, Remaut, H, De Greve, H. | | Deposit date: | 2014-09-10 | | Release date: | 2015-02-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insight in the inhibition of adherence of F4 fimbriae producing enterotoxigenic Escherichia coli by llama single domain antibodies.
Vet. Res., 46, 2015
|
|
2Q6U
 
 | | SeMet-substituted form of NikD | | Descriptor: | BENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, NikD protein | | Authors: | Carrell, C.J, Bruckner, R.C, Venci, D, Zhao, G, Jorns, M.S, Mathews, F.S. | | Deposit date: | 2007-06-05 | | Release date: | 2007-07-31 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | NikD, an Unusual Amino Acid Oxidase Essential for Nikkomycin Biosynthesis: Structures of Closed and Open Forms at 1.15 and 1.90 A Resolution
Structure, 15, 2007
|
|
4WF9
 
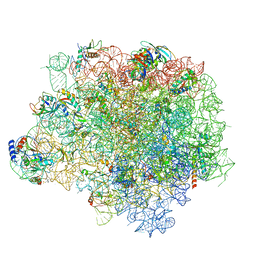 | | The crystal structure of the large ribosomal subunit of Staphylococcus aureus in complex with telithromycin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 23S ribosomal RNA, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Eyal, Z, Matzov, D, Krupkin, M, Wekselman, I, Zimmerman, E, Rozenberg, H, Bashan, A, Yonath, A.E. | | Deposit date: | 2014-09-14 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.427 Å) | | Cite: | Structural insights into species-specific features of the ribosome from the pathogen Staphylococcus aureus.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1U21
 
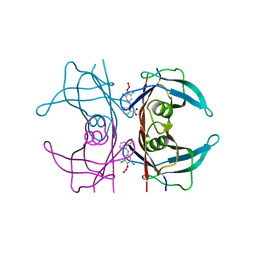 | | transthyretin with tethered inhibitor on one monomer. | | Descriptor: | 2-[(3,5-DICHLORO-4-TRIOXIDANYLPHENYL)AMINO]BENZOIC ACID, Transthyretin | | Authors: | Wiseman, R.L, Johnson, S.M, Kelker, M.S, Foss, T, Wilson, I.A, Kelly, J.W. | | Deposit date: | 2004-07-16 | | Release date: | 2005-06-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Kinetic stabilization of an oligomeric protein by a single ligand binding event
J.Am.Chem.Soc., 127, 2005
|
|
4WRA
 
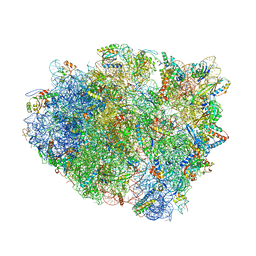 | | Complex of 70S ribosome with tRNA-Tyr and mRNA with A-A mismatch in the first position in the A-site and with antibiotic paromomycin. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rozov, A, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2014-10-23 | | Release date: | 2015-06-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural insights into the translational infidelity mechanism.
Nat Commun, 6, 2015
|
|
8CZC
 
 | | AT from first module of the pikromycin synthase | | Descriptor: | Narbonolide/10-deoxymethynolide synthase PikA1, modules 1 and 2 | | Authors: | Keatinge-Clay, A.T, Dickinson, M.S, Miyazawa, T, McCool, R.S. | | Deposit date: | 2022-05-24 | | Release date: | 2022-06-08 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Priming enzymes from the pikromycin synthase reveal how assembly-line ketosynthases catalyze carbon-carbon chemistry.
Structure, 30, 2022
|
|
4WVC
 
 | | Crystal structure of GH63 mannosylglycerate hydrolase from Thermus thermophilus HB8 in complex with Tris and D-glycerate | | Descriptor: | (2R)-2,3-DIHYDROXYPROPANOIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Miyazaki, T, Ichikawa, M, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2014-11-05 | | Release date: | 2015-03-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and substrate-binding mode of GH63 mannosylglycerate hydrolase from Thermus thermophilus HB8.
J.Struct.Biol., 190, 2015
|
|
6MC6
 
 | |
5KAC
 
 | | Protein Tyrosine Phosphatase 1B P185G mutant, open state | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Choy, M.S, Machado, L.E.S.F, Peti, W, Page, R. | | Deposit date: | 2016-06-01 | | Release date: | 2017-03-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformational Rigidity and Protein Dynamics at Distinct Timescales Regulate PTP1B Activity and Allostery.
Mol. Cell, 65, 2017
|
|
2QYW
 
 | | Crystal structure of mouse vti1b Habc domain | | Descriptor: | Vesicle transport through interaction with t-SNAREs 1B homolog | | Authors: | Miller, S.E, Collins, B.M, McCoy, A.J, Robinson, M.S, Owen, D.J. | | Deposit date: | 2007-08-15 | | Release date: | 2007-11-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A SNARE-adaptor interaction is a new mode of cargo recognition in clathrin-coated vesicles.
Nature, 450, 2007
|
|
7C38
 
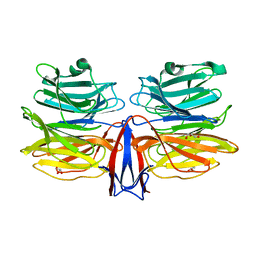 | | Crystal structure of AofleA from Arthrobotrys oligospora in complex with L-fucose | | Descriptor: | AofleA, GLYCEROL, alpha-L-fucopyranose, ... | | Authors: | Liu, M, Cheng, X, Wang, J, Zhang, M, Wang, M. | | Deposit date: | 2020-05-11 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural insights into the fungi-nematodes interaction mediated by fucose-specific lectin AofleA from Arthrobotrys oligospora.
Int.J.Biol.Macromol., 164, 2020
|
|
4O0Q
 
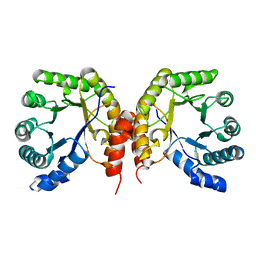 | | Apo structure of a methyltransferase component involved in O-demethylation | | Descriptor: | Dihydropteroate synthase DHPS | | Authors: | Sjuts, H, Dunstan, M.S, Fisher, K, Leys, D. | | Deposit date: | 2013-12-14 | | Release date: | 2015-01-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structures of the methyltransferase component of Desulfitobacterium hafniense DCB-2 O-demethylase shed light on methyltetrahydrofolate formation
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4O1F
 
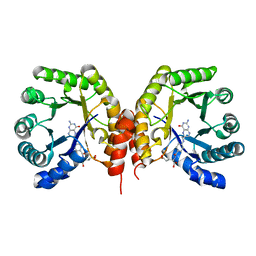 | | Structure of a methyltransferase component in complex with THF involved in O-demethylation | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, Dihydropteroate synthase DHPS | | Authors: | Sjuts, H, Dunstan, M.S, Fisher, K, Leys, D. | | Deposit date: | 2013-12-15 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of the methyltransferase component of Desulfitobacterium hafniense DCB-2 O-demethylase shed light on methyltetrahydrofolate formation
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1U5O
 
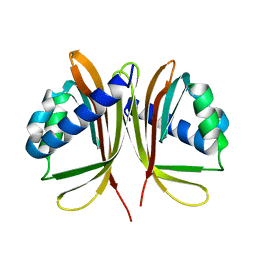 | | Structure of the D23A mutant of the nuclear transport carrier NTF2 | | Descriptor: | Nuclear transport factor 2 | | Authors: | Cushman, I, Bowman, B.R, Sowa, M.E, Lichtarge, O, Quiocho, F.A, Moore, M.S. | | Deposit date: | 2004-07-28 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Computational and biochemical identification of a nuclear pore complex binding site on the nuclear transport carrier NTF2.
J.Mol.Biol., 344, 2004
|
|
4WMG
 
 | | Structure of hen egg-white lysozyme from a microfludic harvesting device using synchrotron radiation (2.5A) | | Descriptor: | Lysozyme C | | Authors: | Lyubimov, A.Y, Murray, T.D, Koehl, A, Uervirojnangkoorn, M, Zeldin, O.B, Cohen, A.E, Soltis, S.M, Baxter, E.M, Brewster, A.S, Sauter, N.K, Brunger, A.T, Berger, J.M. | | Deposit date: | 2014-10-08 | | Release date: | 2015-04-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Capture and X-ray diffraction studies of protein microcrystals in a microfluidic trap array.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WL6
 
 | | Raster-scanning protein crystallography using micro and nano-focused synchrotron beams | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Coquelle, N, Kapp, U, Shilova, A, Weinhausen, B, Burghammer, M, Colletier, J.P. | | Deposit date: | 2014-10-06 | | Release date: | 2015-05-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Raster-scanning serial protein crystallography using micro- and nano-focused synchrotron beams.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
