8IHL
 
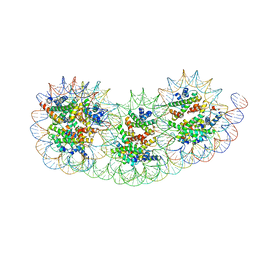 | | Overlapping tri-nucleosome | | Descriptor: | DNA (353-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Nishimura, M, Fujii, T, Tanaka, H, Maehara, K, Nozawa, K, Takizawa, Y, Ohkawa, Y, Kurumizaka, H. | | Deposit date: | 2023-02-23 | | Release date: | 2024-01-17 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (7.64 Å) | | Cite: | Genome-wide mapping and cryo-EM structural analyses of the overlapping tri-nucleosome composed of hexasome-hexasome-octasome moieties.
Commun Biol, 7, 2024
|
|
1ERM
 
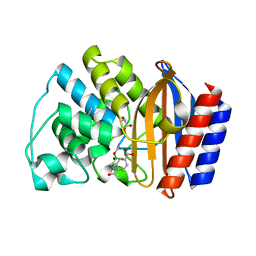 | | X-RAY CRYSTAL STRUCTURE OF TEM-1 BETA LACTAMASE IN COMPLEX WITH A DESIGNED BORONIC ACID INHIBITOR (1R)-1-ACETAMIDO-2-(3-CARBOXYPHENYL)ETHANE BORONIC ACID | | Descriptor: | 1(R)-1-ACETAMIDO-2-(3-CARBOXYPHENYL)ETHYL BORONIC ACID, TEM-1 BETA-LACTAMASE | | Authors: | Ness, S, Martin, R, Kindler, A.M, Paetzel, M, Gold, M, Jones, J.B, Strynadka, N.C.J. | | Deposit date: | 2000-04-06 | | Release date: | 2000-05-10 | | Last modified: | 2012-06-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based design guides the improved efficacy of deacylation transition state analogue inhibitors of TEM-1 beta-Lactamase(,).
Biochemistry, 39, 2000
|
|
6ZMJ
 
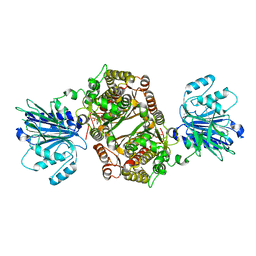 | | Crystal structure of human GFAT-1 R203H | | Descriptor: | GLUCOSE-6-PHOSPHATE, GLUTAMIC ACID, Glutamine--fructose-6-phosphate aminotransferase [isomerizing] 1 | | Authors: | Ruegenberg, S, Mayr, F, Miethe, S, Atanassov, I, Baumann, U, Denzel, M.S. | | Deposit date: | 2020-07-02 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.774 Å) | | Cite: | Protein kinase A controls the hexosamine pathway by tuning the feedback inhibition of GFAT-1.
Nat Commun, 12, 2021
|
|
8IJU
 
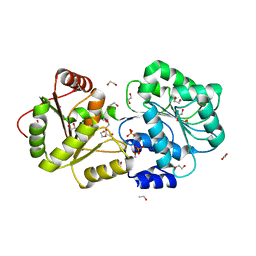 | | ATP-dependent RNA helicase DDX39A (URH49delta41) | | Descriptor: | 1,2-ETHANEDIOL, ATP-dependent RNA helicase DDX39A, PHOSPHATE ION, ... | | Authors: | Mikami, B, Fujita, K, Masuda, S, Kojima, M. | | Deposit date: | 2023-02-28 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural differences between the closely related RNA helicases, UAP56 and URH49, fashion distinct functional apo-complexes.
Nat Commun, 15, 2024
|
|
8WMP
 
 | | Crystal Structure of Mutant HisB from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tiwari, S, Mohini, M, Ahmad, M, Pal, R.K, Biswal, B.K. | | Deposit date: | 2023-10-04 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of Mutant HisB from Mycobacterium tuberculosis
To Be Published
|
|
1EUI
 
 | | ESCHERICHIA COLI URACIL-DNA GLYCOSYLASE COMPLEX WITH URACIL-DNA GLYCOSYLASE INHIBITOR PROTEIN | | Descriptor: | URACIL-DNA GLYCOSYLASE, URACIL-DNA GLYCOSYLASE INHIBITOR PROTEIN | | Authors: | Ravishankar, R, Sagar, M.B, Roy, S, Purnapatre, K, Handa, P, Varshney, U, Vijayan, M. | | Deposit date: | 1998-06-18 | | Release date: | 1999-06-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-ray analysis of a complex of Escherichia coli uracil DNA glycosylase (EcUDG) with a proteinaceous inhibitor. The structure elucidation of a prokaryotic UDG.
Nucleic Acids Res., 26, 1998
|
|
8VQ2
 
 | | HSV1 polymerase ternary complex with dsDNA and compound 44 | | Descriptor: | 2-(4-bromophenyl)-N-(3-methoxy-4-{[(4S)-2-oxo-1,3-oxazolidin-4-yl]methyl}phenyl)acetamide, DNA (5'-D(P*AP*TP*CP*CP*TP*TP*CP*CP*CP*CP*TP*AP*C)-3'), DNA (5'-D(P*TP*GP*GP*TP*AP*GP*GP*GP*GP*AP*AP*GP*GP*AP*T)-3'), ... | | Authors: | Hayes, R.P, Heo, M.R, Plotkin, M. | | Deposit date: | 2024-01-17 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (3.829 Å) | | Cite: | Discovery of Broad-Spectrum Herpes Antiviral Oxazolidinone Amide Derivatives and Their Structure-Activity Relationships.
Acs Med.Chem.Lett., 15, 2024
|
|
4U01
 
 | | HCV NS3/4A serine protease in complex with 6570 | | Descriptor: | (2S,3aS,10Z,11aS,12aR)-2-({8-fluoro-7-methoxy-2-[4-(propan-2-yl)-1,3-thiazol-2-yl]quinolin-4-yl}oxy)-5-methyl-N-[(1-methylcyclopropyl)sulfonyl]-4,14-dioxo-1,2,3,3a,4,5,6,7,8,9,11a,12,13,14-tetradecahydro-12aH-cyclopropa[m]pyrrolo[1,2-c][1,3,6]triazacyclotetradecine-12a-carboxamide, CHLORIDE ION, NS4A protein, ... | | Authors: | Parsy, C.C, Alexandre, F.-R, Brandt, G, Caillet, C, Chaves, D, Derock, M, Gloux, D, Griffon, Y, Lallos, L.B, Leroy, F, Liuzzi, M, Loi, A.-G, Mayes, B, Moulat, L, Moussa, A, Chiara, M, Roques, V, Rosinovsky, E, Seifer, M, Stewart, A, Wang, J, Standring, D, Surleraux, D. | | Deposit date: | 2014-07-11 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery and structural diversity of the hepatitis C virus NS3/4A serine protease inhibitor series leading to clinical candidate IDX320.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
8WS2
 
 | | Crystal Structure of 5'-Deoxy-5'-methylthioadenosine phosphorylase from Aeropyrum pernix complex with 5'-Deoxy-5'-methylthioadenosine | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Iizuka, Y, Kikuchi, M, Yamauchi, T, Tsunoda, M. | | Deposit date: | 2023-10-16 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Crystal Structure of 5'-Deoxy-5'-methylthioadenosine phosphorylase from Aeropyrum pernix complex with 5'-Deoxy-5'-methylthioadenosine
To Be Published
|
|
7DZY
 
 | | Spike protein from SARS-CoV2 with Fab fragment of enhancing antibody 2490 | | Descriptor: | Fab Heavy chain of enhancing antibody 2490, Fab light chain of enhancing antibody 2490, Spike glycoprotein | | Authors: | Liu, Y, Soh, W.T, Li, S, Kishikawa, J, Hirose, M, Kato, T, Standley, D, Okada, M, Arase, H. | | Deposit date: | 2021-01-26 | | Release date: | 2021-06-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | An infectivity-enhancing site on the SARS-CoV-2 spike protein targeted by antibodies.
Cell, 184, 2021
|
|
7DZX
 
 | | Spike protein from SARS-CoV2 with Fab fragment of enhancing antibody 8D2 | | Descriptor: | Fab Heavy chain of enhancing antibody, Fab light chain of enhancing antibody, Spike glycoprotein | | Authors: | Liu, Y, Soh, W.T, Li, S, Kishikawa, J, Hirose, M, Kato, T, Standley, D, Okada, M, Arase, H. | | Deposit date: | 2021-01-26 | | Release date: | 2021-06-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | An infectivity-enhancing site on the SARS-CoV-2 spike protein targeted by antibodies.
Cell, 184, 2021
|
|
7DZW
 
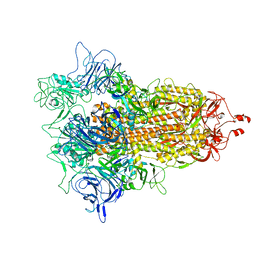 | | Apo spike protein from SARS-CoV2 | | Descriptor: | Spike glycoprotein | | Authors: | Liu, Y, Soh, W.T, Li, S, Kishikawa, J, Hirose, M, Kato, T, Standley, D, Okada, M, Arase, H. | | Deposit date: | 2021-01-26 | | Release date: | 2021-06-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | An infectivity-enhancing site on the SARS-CoV-2 spike protein targeted by antibodies.
Cell, 184, 2021
|
|
1FG4
 
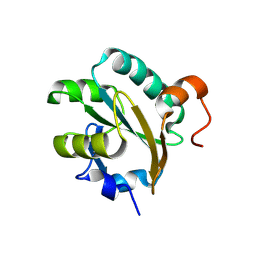 | | STRUCTURE OF TRYPAREDOXIN II | | Descriptor: | TRYPAREDOXIN II | | Authors: | Hofmann, B, Budde, H, Bruns, K, Guerrero, S.A, Kalisz, H.M, Menge, U, Montemartini, M, Nogoceke, E, Steinert, P, Wissing, J.B, Flohe, L, Hecht, H.J. | | Deposit date: | 2000-07-28 | | Release date: | 2001-04-25 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of tryparedoxins revealing interaction with trypanothione.
Biol.Chem., 382, 2001
|
|
8YL9
 
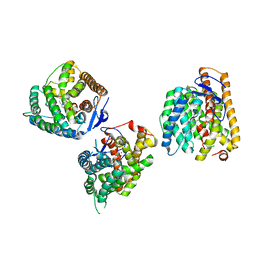 | | Crystal structures of terpene synthases complexed with a substrate mimic | | Descriptor: | MAGNESIUM ION, N-benzyl-N,N-diethylethanaminium, PYROPHOSPHATE, ... | | Authors: | Xu, M, Ma, M. | | Deposit date: | 2024-03-05 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Insights Into the Terpene Cyclization Domains of Two Fungal Sesterterpene Synthases and Enzymatic Engineering for Sesterterpene Diversification.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
6H0K
 
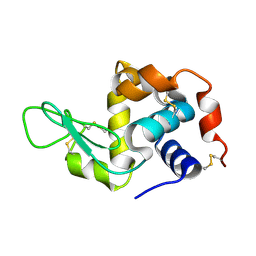 | | Hen egg-white lysozyme structure determined with data from the EuXFEL, the first MHz free electron laser, 7.47 keV photon energy | | Descriptor: | Lysozyme C | | Authors: | Gruenbein, M.L, Gorel, A, Stricker, M, Bean, R, Bielecki, J, Doerner, K, Hartmann, E, Hilpert, M, Kloos, M, Letrun, R, Sztuk-Dambietz, J, Mancuso, A, Meserschmidt, M, Nass-Kovacs, G, Ramilli, M, Roome, C.M, Sato, T, Doak, R.B, Shoeman, R.L, Foucar, L, Colletier, J.P, Barends, T.R.M, Stan, C, Schlichting, I. | | Deposit date: | 2018-07-10 | | Release date: | 2018-09-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Megahertz data collection from protein microcrystals at an X-ray free-electron laser.
Nat Commun, 9, 2018
|
|
6H0L
 
 | | Hen egg-white lysozyme structure determined with data from the EuXFEL, 9.22 keV photon energy | | Descriptor: | Lysozyme C | | Authors: | Gruenbein, M.L, Gorel, A, Stricker, M, Bean, R, Bielecki, J, Doerner, K, Hartmann, E, Hilpert, M, Kloos, M, Letrun, R, Sztuk-Dambietz, J, Mancuso, A, Meserschmidt, M, Nass-Kovacs, G, Ramilli, M, Roome, C.M, Sato, T, Doak, R.B, Shoeman, R.L, Foucar, L, Colletier, J.P, Barends, T.R.M, Stan, C, Schlichting, I. | | Deposit date: | 2018-07-10 | | Release date: | 2018-09-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Megahertz data collection from protein microcrystals at an X-ray free-electron laser.
Nat Commun, 9, 2018
|
|
1FB5
 
 | | LOW RESOLUTION STRUCTURE OF OVINE ORNITHINE TRANSCARBMOYLASE IN THE UNLIGANDED STATE | | Descriptor: | NORVALINE, ORNITHINE TRANSCARBAMOYLASE | | Authors: | Zanotti, G, Battistutta, R, Panzalorto, M, Francescato, P, Bruno, G, De Gregorio, A. | | Deposit date: | 2000-07-14 | | Release date: | 2003-08-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Functional and structural characterization of ovine ornithine transcarbamoylase.
Org.Biomol.Chem., 1, 2003
|
|
8UDU
 
 | | The X-RAY co-crystal structure of human FGFR3 and Compound 17 | | Descriptor: | 3-[(6-chloro-1-cyclopropyl-1H-benzimidazol-5-yl)ethynyl]-1-[(3S,5S)-5-(methoxymethyl)-1-(prop-2-enoyl)pyrrolidin-3-yl]-5-(methylamino)-1H-pyrazole-4-carboxamide, CHLORIDE ION, Fibroblast growth factor receptor 3 | | Authors: | Tyhonas, J.S, Arnold, L.D, Cox, J, Franovic, A, Gardiner, E, Grandinetti, K, Kania, R, Kanouni, T, Lardy, M, Li, C, Martin, E.S, Miller, N, Mohan, A, Murphy, E.A, Perez, M, Soroceanu, L, Timple, N, Uryu, S, Womble, S, Kaldor, S.W. | | Deposit date: | 2023-09-29 | | Release date: | 2024-02-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.737 Å) | | Cite: | Discovery of KIN-3248, An Irreversible, Next Generation FGFR Inhibitor for the Treatment of Advanced Tumors Harboring FGFR2 and/or FGFR3 Gene Alterations.
J.Med.Chem., 67, 2024
|
|
8UDT
 
 | | The X-RAY co-crystal structure of human FGFR3 and KIN-3248 | | Descriptor: | 3-[(1-cyclopropyl-4,6-difluoro-1H-benzimidazol-5-yl)ethynyl]-1-[(3R,5R)-5-(methoxymethyl)-1-propanoylpyrrolidin-3-yl]-5-(methylamino)-1H-pyrazole-4-carboxamide, D-MALATE, Fibroblast growth factor receptor 3 | | Authors: | Tyhonas, J.S, Arnold, L.D, Cox, J, Franovic, A, Gardiner, E, Grandinetti, K, Kania, R, Kanouni, T, Lardy, M, Li, C, Martin, E.S, Miller, N, Mohan, A, Murphy, E.A, Perez, M, Soroceanu, L, Timple, N, Uryu, S, Womble, S, Kaldor, S.W. | | Deposit date: | 2023-09-29 | | Release date: | 2024-02-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.829 Å) | | Cite: | Discovery of KIN-3248, An Irreversible, Next Generation FGFR Inhibitor for the Treatment of Advanced Tumors Harboring FGFR2 and/or FGFR3 Gene Alterations.
J.Med.Chem., 67, 2024
|
|
8YLA
 
 | | Crystal structures of terpene synthases complexed with a substrate mimic | | Descriptor: | MAGNESIUM ION, N-benzyl-N,N-diethylethanaminium, PYROPHOSPHATE, ... | | Authors: | Xu, M, Ma, M. | | Deposit date: | 2024-03-05 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural Insights Into the Terpene Cyclization Domains of Two Fungal Sesterterpene Synthases and Enzymatic Engineering for Sesterterpene Diversification.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
1FDK
 
 | | CARBOXYLIC ESTER HYDROLASE (PLA2-MJ33 INHIBITOR COMPLEX) | | Descriptor: | 1-DECYL-3-TRIFLUORO ETHYL-SN-GLYCERO-2-PHOSPHOMETHANOL, CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | Sundaralingam, M. | | Deposit date: | 1997-09-04 | | Release date: | 1998-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of the complex of bovine pancreatic phospholipase A2 with the inhibitor 1-hexadecyl-3-(trifluoroethyl)-sn-glycero-2-phosphomethanol,.
Biochemistry, 36, 1997
|
|
8XZ2
 
 | | The structural model of a homodimeric D-Ala-D-Ala metallopeptidase, VanX, from vancomycin-resistant bacteria | | Descriptor: | D-alanyl-D-alanine dipeptidase | | Authors: | Konuma, T, Takai, T, Tsuchiya, C, Nishida, M, Hashiba, M, Yamada, Y, Shirai, H, Motoda, Y, Nagadoi, A, Chikaishi, E, Akagi, K, Akashi, S, Yamazaki, T, Akutsu, H, Oe, A, Ikegami, T. | | Deposit date: | 2024-01-20 | | Release date: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Analysis of the homodimeric structure of a D-Ala-D-Ala metallopeptidase, VanX, from vancomycin-resistant bacteria.
Protein Sci., 33, 2024
|
|
1FJS
 
 | | CRYSTAL STRUCTURE OF THE INHIBITOR ZK-807834 (CI-1031) COMPLEXED WITH FACTOR XA | | Descriptor: | CALCIUM ION, CHLORIDE ION, COAGULATION FACTOR XA, ... | | Authors: | Adler, M, Whitlow, M. | | Deposit date: | 2000-08-08 | | Release date: | 2000-10-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Preparation, characterization, and the crystal structure of the inhibitor ZK-807834 (CI-1031) complexed with factor Xa.
Biochemistry, 39, 2000
|
|
8YTN
 
 | | Single-chain Fv antibody of E11 | | Descriptor: | GLYCEROL, Single-chain Fv antibody of E11 | | Authors: | Yoshida, M, Hanazono, Y, Numoto, N, Ito, N, Oda, M. | | Deposit date: | 2024-03-26 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Affinity-matured antibody with a disulfide bond in H-CDR3 loop.
Arch.Biochem.Biophys., 758, 2024
|
|
8YTO
 
 | | Single-chain Fv antibody of E11 complex with NP-glycine | | Descriptor: | 2-[2-(3-nitro-4-oxidanyl-phenyl)ethanoylamino]ethanoic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Yoshida, M, Hanazono, Y, Numoto, N, Ito, N, Oda, M. | | Deposit date: | 2024-03-26 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Affinity-matured antibody with a disulfide bond in H-CDR3 loop.
Arch.Biochem.Biophys., 758, 2024
|
|
