3WYF
 
 | | Crystal structure of Xpo1p-Yrb2p-Gsp1p-GTP complex | | Descriptor: | Exportin-1, GUANOSINE-5'-TRIPHOSPHATE, Gsp1p, ... | | Authors: | Koyama, M, Shirai, N, Matsuura, Y. | | Deposit date: | 2014-08-26 | | Release date: | 2014-11-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural insights into how yrb2p accelerates the assembly of the xpo1p nuclear export complex
Cell Rep, 9, 2014
|
|
2LL5
 
 | | Cyclo-TC1 Trp-cage | | Descriptor: | Cyclo-TC1 | | Authors: | Lin, J.C, Scian, M, Andersen, N.H. | | Deposit date: | 2011-10-26 | | Release date: | 2012-07-18 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Crystal and NMR structures of a Trp-cage mini-protein benchmark for computational fold prediction.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4EZA
 
 | | Crystal structure of the atypical phosphoinositide (aPI) binding domain of IQGAP2 | | Descriptor: | Ras GTPase-activating-like protein IQGAP2 | | Authors: | Van Aalten, D.M.F, Dixon, M.J, Gray, A, Schenning, M, Agacan, M, Leslie, N.R, Downes, C.P, Batty, I.H, Nedyalkova, L, Tempel, W, Tong, Y, Zhong, N, Crombet, L, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-05-02 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | IQGAP Proteins Reveal an Atypical Phosphoinositide (aPI) Binding Domain with a Pseudo C2 Domain Fold.
J.Biol.Chem., 287, 2012
|
|
2L1N
 
 | | Solution NMR structure of the protein YP_399305.1 | | Descriptor: | Uncharacterized protein | | Authors: | Mohanty, B, Serrano, P, Geralt, M, Horst, R, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2010-07-30 | | Release date: | 2010-08-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the protein YP_399305.1
To be Published
|
|
2LME
 
 | | Solid-state NMR structure of the membrane anchor domain of the trimeric autotransporter YadA | | Descriptor: | Adhesin yadA | | Authors: | Shahid, S.A, Bardiaux, B, Franks, W.T, Habeck, M, Linke, D, van Rossum, B. | | Deposit date: | 2011-11-30 | | Release date: | 2012-11-07 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Membrane-protein structure determination by solid-state NMR spectroscopy of microcrystals.
Nat.Methods, 9, 2012
|
|
2L4F
 
 | |
3ZMH
 
 | | Structure of E.coli rhomboid protease GlpG in complex with monobactam L62 | | Descriptor: | CHLORIDE ION, CYCLOPENTYL 2-OXO-4-PHENYLAZETIDINE-1-CARBOXYLATE, RHOMBOID PROTEASE GLPG, ... | | Authors: | Vinothkumar, K.R, Pierrat, O.A, Large, J.M, Freeman, M. | | Deposit date: | 2013-02-11 | | Release date: | 2013-05-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of rhomboid protease in complex with beta-lactam inhibitors defines the S2' cavity.
Structure, 21, 2013
|
|
3X2J
 
 | | X-ray structure of PcCel45A D114N apo form at 95K. | | Descriptor: | 3-methylpentane-1,5-diol, Endoglucanase V-like protein | | Authors: | Nakamura, A, Ishida, T, Samejima, M, Igarashi, K. | | Deposit date: | 2014-12-22 | | Release date: | 2015-10-07 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.301 Å) | | Cite: | "Newton's cradle" proton relay with amide-imidic acid tautomerization in inverting cellulase visualized by neutron crystallography.
Sci Adv, 1, 2015
|
|
2PJ2
 
 | |
2LF3
 
 | | Solution NMR structure of HopPmaL_281_385 from Pseudomonas syringae pv. maculicola str. ES4326, Midwest Center for Structural Genomics target APC40104.5 and Northeast Structural Genomics Consortium target PsT2A | | Descriptor: | Effector protein hopAB3 | | Authors: | Wu, B, Yee, A, Houliston, S, Semesi, A, Garcia, M, Singer, A.U, Savchenko, A, Montelione, G.T, Joachimiak, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Midwest Center for Structural Genomics (MCSG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2011-06-28 | | Release date: | 2011-07-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of HopPmaL Reveals the Presence of a Second Adaptor Domain Common to the HopAB Family of Pseudomonas syringae Type III Effectors.
Biochemistry, 51, 2012
|
|
2L9Q
 
 | |
3X36
 
 | | Crystal structure of the human vitamin D receptor ligand binding domain complexed with 7,8-cis-1a,25-Dihydroxy-19-norvitamin D3 | | Descriptor: | (1R,3R)-5-[(2Z)-2-[(1R,3aS,7aR)-7a-methyl-1-[(2R)-6-methyl-6-oxidanyl-heptan-2-yl]-2,3,3a,5,6,7-hexahydro-1H-inden-4-ylidene]ethylidene]cyclohexane-1,3-diol, Vitamin D3 receptor | | Authors: | Takimoto-Kamimura, M, Kakuda, S. | | Deposit date: | 2015-01-16 | | Release date: | 2016-01-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Revisiting the 7,8-cis-vitamin D3 derivatives: synthesis, evaluating the biological activity, and study of the binding configuration
To be Published
|
|
2LIT
 
 | | NMR Solution Structure of Yeast Iso-1-cytochrome c Mutant P71H in reduced states | | Descriptor: | Cytochrome c iso-1, HEME C | | Authors: | Lan, W, Wang, Z, Yang, Z, Zhu, J, Ying, T, Jiang, X, Zhang, X, Wu, H, Liu, M, Tan, X, Cao, C, Huang, Z.X. | | Deposit date: | 2011-08-31 | | Release date: | 2011-12-07 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Conformational toggling of yeast iso-1-cytochrome C in the oxidized and reduced States.
Plos One, 6, 2011
|
|
3X3Z
 
 | | Copper amine oxidase from Arthrobacter globiformis: Aminoresorcinol form produced by anaerobic reduction with ethylamine hydrochloride | | Descriptor: | CHLORIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Okajima, T, Nakanishi, S, Murakawa, T, Kataoka, M, Hayashi, H, Hamaguchi, A, Nakai, T, Kawano, Y, Yamaguchi, H, Tanizawa, K. | | Deposit date: | 2015-03-10 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Probing the Catalytic Mechanism of Copper Amine Oxidase from Arthrobacter globiformis with Halide Ions.
J.Biol.Chem., 290, 2015
|
|
3X0X
 
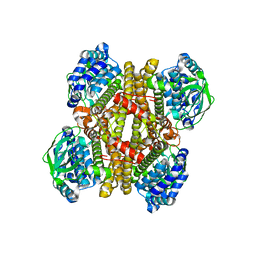 | | Crystal structure of apo-DszC from Rhodococcus erythropolis D-1 | | Descriptor: | DszC | | Authors: | Guan, L.J, Lee, W.C, Wang, S.P, Ohtsuka, J, Tanokura, M. | | Deposit date: | 2014-10-23 | | Release date: | 2015-02-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal structures of apo-DszC and FMN-bound DszC from Rhodococcus erythropolis D-1.
Febs J., 282, 2015
|
|
4F3Q
 
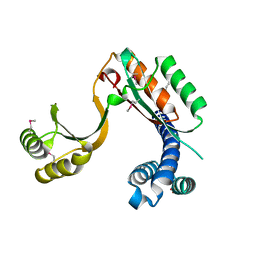 | | Structure of a YebC family protein (CBU_1566) from Coxiella burnetii | | Descriptor: | SULFATE ION, Transcriptional regulatory protein CBU_1566 | | Authors: | Franklin, M.C, Cheung, J, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2012-05-09 | | Release date: | 2012-06-27 | | Last modified: | 2016-02-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
3X2G
 
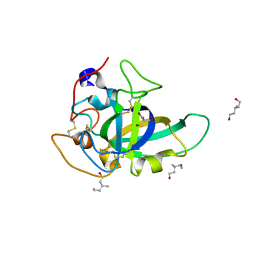 | | X-ray structure of PcCel45A N92D apo form at 100K | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-methylpentane-1,5-diol, Endoglucanase V-like protein | | Authors: | Nakamura, A, Ishida, T, Samejima, M, Igarashi, K. | | Deposit date: | 2014-12-22 | | Release date: | 2015-10-07 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | "Newton's cradle" proton relay with amide-imidic acid tautomerization in inverting cellulase visualized by neutron crystallography.
Sci Adv, 1, 2015
|
|
2LMI
 
 | |
2LNZ
 
 | |
8A8L
 
 | | Crystal structure of a staphylococcal orthologue of CYP134A1 (CYPX) in complex with a heme-coordinated fragment | | Descriptor: | 6-methoxy-2,3,4,9-tetrahydro-1H-pyrido[3,4-b]indole, Cytochrome P450 protein, GLYCEROL, ... | | Authors: | Snee, M, Katariya, M, Levy, C. | | Deposit date: | 2022-06-23 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure of a staphylococcal orthologue of CYP134A1 (CYPX) in complex with a heme-coordinated fragment
To Be Published
|
|
2LSH
 
 | |
4Y0S
 
 | | Goat beta-lactoglobulin complex with pramocaine (GLG-PRM) | | Descriptor: | Beta-lactoglobulin, Pramocaine, SULFATE ION | | Authors: | Loch, J.I, Bonarek, P, Polit, A, Jablonski, M, Czub, M, Ye, X, Lewinski, K. | | Deposit date: | 2015-02-06 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | beta-Lactoglobulin interactions with local anaesthetic drugs - Crystallographic and calorimetric studies.
Int.J.Biol.Macromol., 80, 2015
|
|
2PJB
 
 | | CRYSTAL STRUCTURE OF ACTIVATED PORCINE PANCREATIC CARBOXYPEPTIDASE B 2-(3-Aminomethyl-phenyl)-3-{[1-((S)-2-benzyloxycarbonylamino-3-phenyl-propane-1-sulfonylamino)-2-methyl-propyl]-hydroxy-phosphinoyl}-propionic acid COMPLEX | | Descriptor: | (5S,9R,10R,12S)-12-[3-(AMINOMETHYL)PHENYL]-5-BENZYL-10-HYDROXY-9-ISOPROPYL-3-OXO-1-PHENYL-2-OXA-7-THIA-4,8-DIAZA-10-PHOSPHATRIDECAN-13-OIC ACID 7,7,10-TRIOXIDE, Carboxypeptidase B, ZINC ION | | Authors: | Adler, M, Whitlow, M. | | Deposit date: | 2007-04-15 | | Release date: | 2008-01-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of potent selective peptide mimetics bound to carboxypeptidase B.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3WZ6
 
 | | Endothiapepsin in complex with Gewald reaction-derived inhibitor (5) | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kuhnert, M, Steuber, H, Diederich, W.E. | | Deposit date: | 2014-09-19 | | Release date: | 2015-08-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.404 Å) | | Cite: | Tracing binding modes in hit-to-lead optimization: chameleon-like poses of aspartic protease inhibitors
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
3ZM9
 
 | | The mechanism of allosteric coupling in choline kinase a1 revealed by a rationally designed inhibitor | | Descriptor: | 1-(4-(4-(4-((6-amino-9H-purin-9-yl)methyl)phenyl)butyl)benzyl)-4- (dimethylamino)pyridinium, CHOLINE KINASE ALPHA | | Authors: | Sahun-Roncero, M, Rubio-Ruiz, B, Saladino, G, Conejo-Garcia, A, Espinosa, A, Velazquez-Campoy, A, Gervasio, F.L, Entrena, A, Hurtado-Guerrero, R. | | Deposit date: | 2013-02-06 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Mechanism of Allosteric Coupling in Choline Kinase A1 Revealed by a Rationally Designed Inhibitor
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
