4J59
 
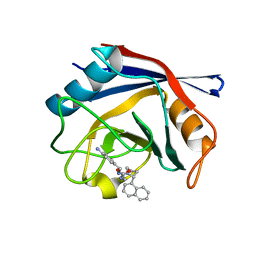 | | Human Cyclophilin D Complexed with an Inhibitor | | Descriptor: | 1-(4-aminobenzyl)-3-{2-[(2R)-2-(naphthalen-1-yl)pyrrolidin-1-yl]-2-oxoethyl}urea, Peptidyl-prolyl cis-trans isomerase F, mitochondrial | | Authors: | Gelin, M, Colliandre, L, Bessin, Y, Guichou, J.F. | | Deposit date: | 2013-02-08 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Fragment-based discovery of a new family of non-peptidic small-molecule cyclophilin inhibitors with potent antiviral activities.
Nat Commun, 7, 2016
|
|
1KOG
 
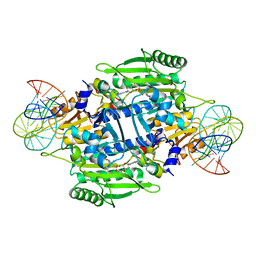 | | Crystal structure of E. coli threonyl-tRNA synthetase interacting with the essential domain of its mRNA operator | | Descriptor: | 5'-O-(N-(L-THREONYL)-SULFAMOYL)ADENOSINE, Threonyl-tRNA synthetase, Threonyl-tRNA synthetase mRNA, ... | | Authors: | Torres-Larrios, A, Dock-Bregeon, A.C, Romby, P, Rees, B, Sankaranarayanan, R, Caillet, J, Springer, M, Ehresmann, C, Ehresmann, B, Moras, D. | | Deposit date: | 2001-12-20 | | Release date: | 2002-04-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis of translational control by Escherichia coli threonyl tRNA synthetase.
Nat.Struct.Biol., 9, 2002
|
|
3ROZ
 
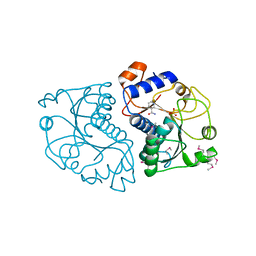 | | Crystal Structure of Mouse Apolipoprotein A-I Binding Protein in Complex with Nicotinamide | | Descriptor: | Apolipoprotein A-I-binding protein, NICOTINAMIDE, SULFATE ION | | Authors: | Shumilin, I.A, Jha, K.N, Cymborowski, M, Herr, J.C, Minor, W. | | Deposit date: | 2011-04-26 | | Release date: | 2012-07-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
6GQC
 
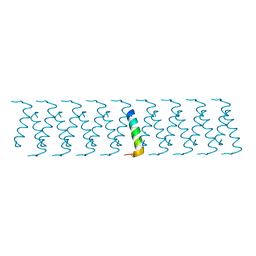 | |
4QAL
 
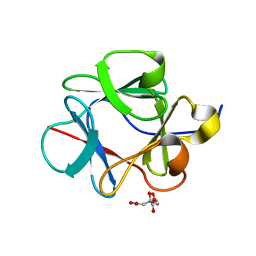 | |
4J67
 
 | | Crystal structure of Ribonuclease A soaked in 50% 1,6-Hexanediol: One of twelve in MSCS set | | Descriptor: | HEXANE-1,6-DIOL, Ribonuclease pancreatic, SULFATE ION | | Authors: | Kearney, B.M, Dechene, M, Swartz, P.D, Mattos, C. | | Deposit date: | 2013-02-11 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | DRoP: A program for analysis of water structure on protein surfaces
to be published
|
|
3RQ8
 
 | | Crystal Structure of ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Bacillus subtilis soaked with P1,P5-Di(adenosine-5') pentaphosphate | | Descriptor: | ADP/ATP-dependent NAD(P)H-hydrate dehydratase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION | | Authors: | Shumilin, I.A, Cymborowski, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-27 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
6U4P
 
 | | Structure-based discovery of a novel small-molecule inhibitor of methicillin-resistant S. aureus | | Descriptor: | Alpha-hemolysin, SULFATE ION, fos-choline-14 | | Authors: | Liu, J, Kozhaya, L, Torres, V.J, Unutmaz, D, Lu, M. | | Deposit date: | 2019-08-26 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structure-based discovery of a small-molecule inhibitor of methicillin-resistantStaphylococcus aureusvirulence.
J.Biol.Chem., 295, 2020
|
|
6KEX
 
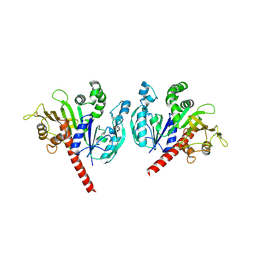 | |
3DMI
 
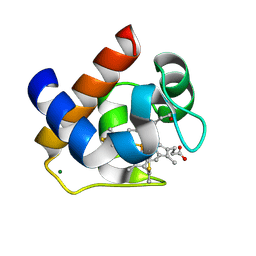 | | Crystallization and Structural Analysis of Cytochrome c6 from the Diatom Phaeodactylum tricornutum at 1.5 A resolution | | Descriptor: | HEME C, MAGNESIUM ION, cytochrome c6 | | Authors: | Akazaki, H, Kawai, F, Hosokawa, M, Hama, T, Hirano, T, Lim, B.-K, Sakurai, N, Hakamata, W, Park, S.-Y, Nishio, T, Oku, T. | | Deposit date: | 2008-07-01 | | Release date: | 2009-03-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallization and structural analysis of cytochrome c(6) from the diatom Phaeodactylum tricornutum at 1.5 A resolution.
Biosci.Biotechnol.Biochem., 73, 2009
|
|
5A4X
 
 | |
3RS9
 
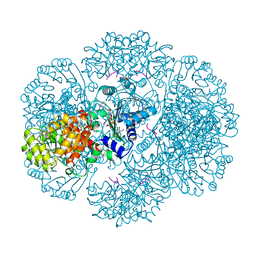 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima in complex with P1,P3-Di(adenosine-5') triphosphate | | Descriptor: | BIS(ADENOSINE)-5'-TRIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-05-02 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
6GRH
 
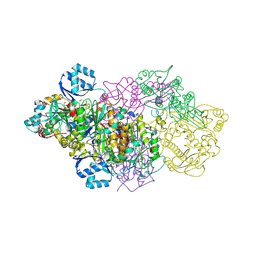 | | E. coli Microcin synthetase McbBCD complex with truncated pro-MccB17 bound | | Descriptor: | 1,2-ETHANEDIOL, Bacteriocin microcin B17, CHLORIDE ION, ... | | Authors: | Ghilarov, D, Stevenson, C.E.M, Travin, D.Y, Piskunova, J, Serebryakova, M, Maxwell, A, Lawson, D.M, Severinov, K. | | Deposit date: | 2018-06-11 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Architecture of Microcin B17 Synthetase: An Octameric Protein Complex Converting a Ribosomally Synthesized Peptide into a DNA Gyrase Poison.
Mol. Cell, 73, 2019
|
|
3RSQ
 
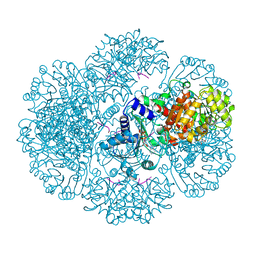 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima soaked with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, BETA-6-HYDROXY-1,4,5,6-TETRHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, POTASSIUM ION, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-05-02 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.054 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
4QHF
 
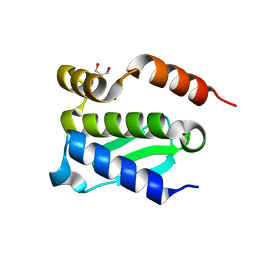 | | Crystal structure of Methanocaldococcus jannaschii monomeric selecase | | Descriptor: | GLYCEROL, NICKEL (II) ION, Uncharacterized protein MJ1213 | | Authors: | Lopez-pelegrin, M, Cerda-costa, N, Cintas-pedrola, A, Herranz-trillo, F, Bernado, P, Peinado, J.R, Arolas, J.L, Gomis-ruth, F.X. | | Deposit date: | 2014-05-28 | | Release date: | 2014-07-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Multiple stable conformations account for reversible concentration-dependent oligomerization and autoinhibition of a metamorphic metallopeptidase
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3RTG
 
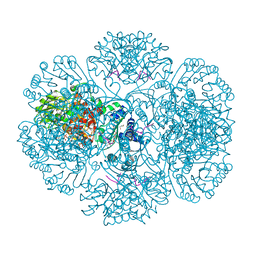 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima soaked with Coenzyme A and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, COENZYME A, MAGNESIUM ION, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-05-03 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
6GSQ
 
 | | Oxidised copper nitrite reductase from Achromobacter cycloclastes determined by serial femtosecond rotation crystallography | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, SULFATE ION | | Authors: | Halsted, T.P, Yamashita, K, Gopalasingam, C.C, Shenoy, R.T, Hirata, K, Ago, H, Ueno, G, Eady, R.R, Antonyuk, S.V, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2018-06-15 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Catalytically important damage-free structures of a copper nitrite reductase obtained by femtosecond X-ray laser and room-temperature neutron crystallography.
Iucrj, 6, 2019
|
|
4QCA
 
 | | Crystal structure of Vaccinia virus uracil-DNA glycosylase mutant R167AD4 | | Descriptor: | CHLORIDE ION, GLYCEROL, POTASSIUM ION, ... | | Authors: | Sartmatova, D, Nash, T, Schormann, N, Nuth, M, Ricciardi, R, Banerjee, S, Chattopadhyay, D. | | Deposit date: | 2014-05-09 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallization and preliminary X-ray diffraction analysis of three recombinant mutants of Vaccinia virus uracil DNA glycosylase.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4J8R
 
 | | Structure of an octapeptide repeat of the prion protein bound to the POM2 Fab antibody fragment | | Descriptor: | Heavy chain of POM2 Fab, Light chain of POM2 Fab, Major prion protein | | Authors: | Swayampakula, M, Baral, P.K, Kav, N.N.V, Aguzzi, A, James, M.N.G. | | Deposit date: | 2013-02-14 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | The crystal structure of an octapeptide repeat of the Prion protein in complex with a Fab fragment of the POM2 antibody.
Protein Sci., 22, 2013
|
|
6KGA
 
 | |
3RTO
 
 | | Acoustically mounted porcine insulin microcrystals yield an X-ray SAD structure | | Descriptor: | Insulin, ZINC ION | | Authors: | Soares, A.S, Engel, M.A, Stearns, R, Datwani, S, Olechno, J, Ellson, R, Skinner, J.M, Allaire, M, Orville, A.M. | | Deposit date: | 2011-05-03 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Acoustically Mounted Microcrystals Yield High-Resolution X-ray Structures.
Biochemistry, 50, 2011
|
|
6GT6
 
 | | Crystal structure of recombinant coagulation factor beta-XIIa | | Descriptor: | CYSTEINE, Coagulation factor XII, GLYCEROL, ... | | Authors: | Pathak, M, Emsley, J. | | Deposit date: | 2018-06-15 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Crystal structures of the recombinant beta-factor XIIa protease with bound Thr-Arg and Pro-Arg substrate mimetics.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
1KTO
 
 | |
1KU2
 
 | | Crystal Structure of Thermus aquaticus RNA Polymerase Sigma Subunit Fragment Containing Regions 1.2 to 3.1 | | Descriptor: | SULFATE ION, sigma factor sigA | | Authors: | Campbell, E.A, Muzzin, O, Chlenov, M, Sun, J.L, Olson, C.A, Weinman, O, Trester-Zedlitz, M.L, Darst, S.A. | | Deposit date: | 2002-01-21 | | Release date: | 2002-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the bacterial RNA polymerase promoter specificity sigma subunit.
Mol.Cell, 9, 2002
|
|
6KHJ
 
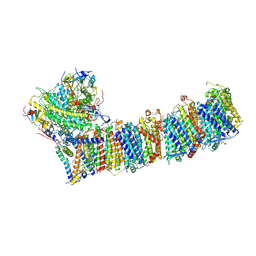 | | Supercomplex for electron transfer | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Pan, X, Cao, D, Xie, F, Zhang, X, Li, M. | | Deposit date: | 2019-07-15 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for electron transport mechanism of complex I-like photosynthetic NAD(P)H dehydrogenase.
Nat Commun, 11, 2020
|
|
