5QUN
 
 | | Structure of unliganded HumRadA1.6 | | Descriptor: | PHOSPHATE ION, RadA | | Authors: | Marsh, M, Hyvonen, M. | | Deposit date: | 2020-01-27 | | Release date: | 2021-03-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Optimisation of crystal forms for structure-guided drug discovery
To be published
|
|
8DL7
 
 | | Cryo-EM structure of human ferroportin/slc40 bound to minihepcidin PR73 in nanodisc | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 11F9 heavy-chain, 11F9 light-chain, ... | | Authors: | Shen, J, Wilbon, A.S, Pan, Y, Zhou, M. | | Deposit date: | 2022-07-07 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of ferroportin inhibition by minihepcidin PR73.
Plos Biol., 21, 2023
|
|
5QUM
 
 | | Structure of unliganded HumRadA1.4 | | Descriptor: | RadA | | Authors: | Marsh, M, Hyvonen, M. | | Deposit date: | 2020-01-27 | | Release date: | 2021-03-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Optimisation of crystal forms for structure-guided drug discovery
To be published
|
|
5QUL
 
 | | Structure of unliganded HumRadA1.3 | | Descriptor: | PHOSPHATE ION, RadA | | Authors: | Marsh, M, Hyvonen, M. | | Deposit date: | 2020-01-27 | | Release date: | 2021-03-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.278 Å) | | Cite: | Optimisation of crystal forms for structure-guided drug discovery
To be published
|
|
5QUT
 
 | |
5QUS
 
 | | HumRadA1 soaked with 5 mM indazole in 10% DMSO | | Descriptor: | 1H-indazole, DIMETHYL SULFOXIDE, PHOSPHATE ION, ... | | Authors: | Marsh, M, Hyvonen, M. | | Deposit date: | 2020-01-27 | | Release date: | 2021-03-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.247 Å) | | Cite: | Optimisation of crystal forms for structure-guided drug discovery
To be published
|
|
8EAI
 
 | | SsoMCM hexamer bound to Mg/ADP-BeFx and 16-mer oligo-dT. Class 2 | | Descriptor: | 16-mer oligo-dT, MAGNESIUM ION, Minichromosome maintenance protein MCM, ... | | Authors: | Meagher, M, Myasnikov, A, Enemark, E.J. | | Deposit date: | 2022-08-29 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Two Distinct Modes of DNA Binding by an MCM Helicase Enable DNA Translocation.
Int J Mol Sci, 23, 2022
|
|
5QUU
 
 | | HumRadA1 as soaking control with 10% DMSO | | Descriptor: | DIMETHYL SULFOXIDE, PHOSPHATE ION, RadA | | Authors: | Marsh, M, Hyvonen, M. | | Deposit date: | 2020-01-27 | | Release date: | 2021-03-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.248 Å) | | Cite: | Optimisation of crystal forms for structure-guided drug discovery
To be published
|
|
8DL8
 
 | | Cryo-EM structure of human ferroportin/slc40 bound to Co2+ in nanodisc | | Descriptor: | 11F9 heavy-chain, 11F9 light-chain, COBALT (II) ION, ... | | Authors: | Shen, J, Wilbon, A.S, Pan, Y, Zhou, M. | | Deposit date: | 2022-07-07 | | Release date: | 2022-12-07 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of ferroportin inhibition by minihepcidin PR73.
Plos Biol., 21, 2023
|
|
5QUR
 
 | |
5K64
 
 | | Crystal structure of VEGF binding IgG1-Fc (Fcab 448) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Humm, A, Lobner, E, Kitzmuller, M, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | Deposit date: | 2016-05-24 | | Release date: | 2017-09-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Two-faced Fcab prevents polymerization with VEGF and reveals thermodynamics and the 2.15 angstrom crystal structure of the complex.
MAbs, 9, 2017
|
|
8EAK
 
 | | SsoMCM hexamer bound to Mg/ADP-BeFx and 46-mer DNA strand. Class 2 | | Descriptor: | 46-mer DNA, MAGNESIUM ION, Minichromosome maintenance protein MCM, ... | | Authors: | Meagher, M, Myasnikov, A, Enemark, E.J. | | Deposit date: | 2022-08-29 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Two Distinct Modes of DNA Binding by an MCM Helicase Enable DNA Translocation.
Int J Mol Sci, 23, 2022
|
|
1CCH
 
 | |
5T4L
 
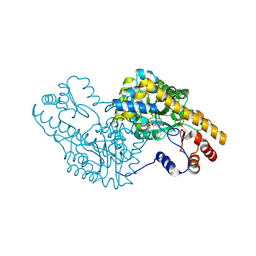 | | PLP and GABA Trigger GabR-Mediated Transcription Regulation in Bacillus subsidies via External Aldimine Formation | | Descriptor: | (4R)-4-amino-6-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}hexanoic acid, Aspartate aminotransferase | | Authors: | Wu, R, Sanishvili, R, Belitsky, B.R, Juncosa, J.I, Le, H.V, Lehrer, H.J.S, Farley, M, Silverman, R.B, Petsko, G.A, Ringe, D, Liu, D. | | Deposit date: | 2016-08-29 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | PLP and GABA trigger GabR-mediated transcription regulation in Bacillus subtilis via external aldimine formation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8EAM
 
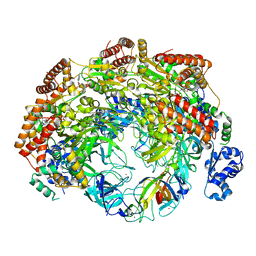 | | SsoMCM hexamer bound to Mg/ADP-BeFx and DNA. Class 2. Merged particles from datasets with 3 different DNA entities | | Descriptor: | DNA, MAGNESIUM ION, Minichromosome maintenance protein MCM, ... | | Authors: | Meagher, M, Myasnikov, A, Enemark, E.J. | | Deposit date: | 2022-08-29 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Two Distinct Modes of DNA Binding by an MCM Helicase Enable DNA Translocation.
Int J Mol Sci, 23, 2022
|
|
5K9S
 
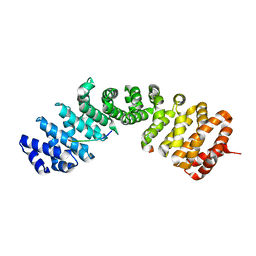 | | Importin-alpha in complex with HNF1-beta peptide | | Descriptor: | HNF1 beta A splice variant 2, Importin subunit alpha-1 | | Authors: | Wiedmann, M.M, Aibara, S, Spring, D.R, Stewart, M, Brenton, J. | | Deposit date: | 2016-06-01 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Importin-alpha in complex with HNF1-beta peptide
To Be Published
|
|
5GO7
 
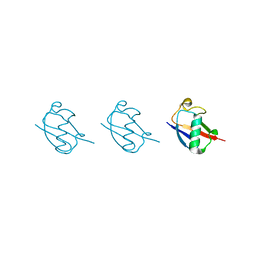 | | Linear tri-ubiquitin | | Descriptor: | D-ubiquitin, Ubiquitin | | Authors: | Gao, S, Pan, M, Zheng, Y. | | Deposit date: | 2016-07-26 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Monomer/Oligomer Quasi-Racemic Protein Crystallography
J.Am.Chem.Soc., 138, 2016
|
|
7R2V
 
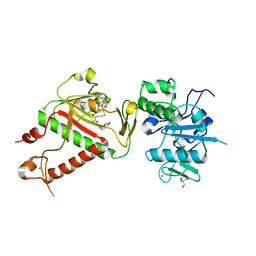 | | Structure of nsp14 from SARS-CoV-2 in complex with SAH | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, Proofreading exoribonuclease nsp14, ... | | Authors: | Czarna, A, Plewka, J, Kresik, L, Matsuda, A, Abdulkarim, K, Robinson, C, OByrne, S, Cunningham, F, Georgiou, I, Pachota, M, Popowicz, G.M, Wyatt, P.G, Dubin, G, Pyrc, K. | | Deposit date: | 2022-02-06 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Refolding of lid subdomain of SARS-CoV-2 nsp14 upon nsp10 interaction releases exonuclease activity.
Structure, 30, 2022
|
|
5GOC
 
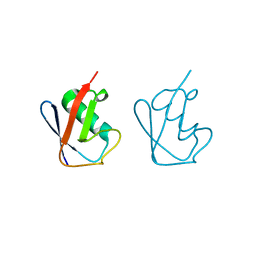 | | Lys11-linked diubiquitin | | Descriptor: | D-ubiquitin, SODIUM ION, Ubiquitin | | Authors: | Gao, S, Pan, M, Zheng, Y. | | Deposit date: | 2016-07-26 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.733 Å) | | Cite: | Monomer/Oligomer Quasi-Racemic Protein Crystallography
J.Am.Chem.Soc., 138, 2016
|
|
2O1O
 
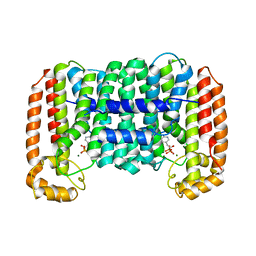 | | Cryptosporidium parvum putative polyprenyl pyrophosphate synthase (cgd4_2550) in complex with risedronate. | | Descriptor: | 1-HYDROXY-2-(3-PYRIDINYL)ETHYLIDENE BIS-PHOSPHONIC ACID, MAGNESIUM ION, Putative farnesyl pyrophosphate synthase | | Authors: | Chruszcz, M, Artz, J.D, Dong, A, Dunford, J, Lew, J, Zhao, Y, Kozieradski, I, Kavanaugh, K.L, Oppermann, U, Sundstrom, M, Weigelt, J, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Hui, R, Minor, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-11-29 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Targeting a uniquely nonspecific prenyl synthase with bisphosphonates to combat cryptosporidiosis
Chem.Biol., 15, 2008
|
|
8EAJ
 
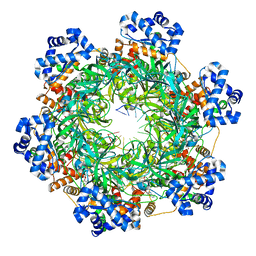 | | SsoMCM hexamer bound to Mg/ADP-BeFx and 46-mer DNA strand. Class 1 | | Descriptor: | 46-mer DNA strand, MAGNESIUM ION, Minichromosome maintenance protein MCM, ... | | Authors: | Meagher, M, Myasnikov, A, Enemark, E.J. | | Deposit date: | 2022-08-29 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Two Distinct Modes of DNA Binding by an MCM Helicase Enable DNA Translocation.
Int J Mol Sci, 23, 2022
|
|
5GOI
 
 | | Lys48-linked di-ubiquitin | | Descriptor: | D-ubiquitin, Ubiquitin | | Authors: | Gao, S, Pan, M, Zheng, Y, Liu, L. | | Deposit date: | 2016-07-27 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.594 Å) | | Cite: | Monomer/Oligomer Quasi-Racemic Protein Crystallography
J.Am.Chem.Soc., 138, 2016
|
|
8EAH
 
 | | SsoMCM hexamer bound to Mg/ADP-BeFx and 16-mer oligo-dT. Class 1 | | Descriptor: | 16-mer oligo-dT, MAGNESIUM ION, Minichromosome maintenance protein MCM, ... | | Authors: | Meagher, M, Myasnikov, A, Enemark, E.J. | | Deposit date: | 2022-08-29 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Two Distinct Modes of DNA Binding by an MCM Helicase Enable DNA Translocation.
Int J Mol Sci, 23, 2022
|
|
1BTV
 
 | | STRUCTURE OF BET V 1, NMR, 20 STRUCTURES | | Descriptor: | BET V 1 | | Authors: | Osmark, P, Poulsen, F.M, Gajhede, M, Larsen, J.N, Spangfort, M.D. | | Deposit date: | 1997-01-30 | | Release date: | 1997-08-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | X-ray and NMR structure of Bet v 1, the origin of birch pollen allergy.
Nat.Struct.Biol., 3, 1996
|
|
8EAG
 
 | | SsoMCM hexamer bound to Mg/ADP-BeFx and 12-mer oligo-dT. Class 2 | | Descriptor: | 12-mer oligo-dT, MAGNESIUM ION, Minichromosome maintenance protein MCM, ... | | Authors: | Meagher, M, Myasnikov, A, Enemark, E.J. | | Deposit date: | 2022-08-29 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Two Distinct Modes of DNA Binding by an MCM Helicase Enable DNA Translocation.
Int J Mol Sci, 23, 2022
|
|
