5JH7
 
 | | Tubulin-Eribulin complex | | Descriptor: | (1S,3S,6S,9S,12S,14R,16R,18S,20R,21R,22S,26R,29S,31R,32S,33R,35R,36S)-20-[(2S)-3-amino-2-hydroxypropyl]-21-methoxy-14-methyl-8,15-dimethylidene-2,19,30,34,37,39,40,41-octaoxanonacyclo[24.9.2.1~3,32~.1~3,33~.1~6,9~.1~12,16~.0~18,22~.0~29,36~.0~31,35~]hentetracontan-24-one (non-preferred name), 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Doodhi, H, Prota, A.E, Rodriguez-Garcia, R, Xiao, H, Custar, D.W, Bargsten, K, Katrukha, E.A, Hilbert, M, Hua, S, Jiang, K, Grigoriev, I, Yang, C.-P.H, Cox, D, Band Horwitz, S, Kapitein, L.C, Akhmanova, A, Steinmetz, M.O. | | Deposit date: | 2016-04-20 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Termination of Protofilament Elongation by Eribulin Induces Lattice Defects that Promote Microtubule Catastrophes.
Curr.Biol., 26, 2016
|
|
6YRT
 
 | |
5GZK
 
 | | Endo-beta-1,2-glucanase from Chitinophaga pinensis - sophorotriose and glucose complex | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, ... | | Authors: | Abe, K, Nakajima, M, Arakawa, T, Fushinobu, S, Taguchi, H. | | Deposit date: | 2016-09-28 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and structural analyses of a bacterial endo-beta-1,2-glucanase reveal a new glycoside hydrolase family
J. Biol. Chem., 292, 2017
|
|
2LZT
 
 | |
6YS0
 
 | |
6YRM
 
 | |
2XQB
 
 | | Crystal Structure of anti-IL-15 Antibody in Complex with human IL-15 | | Descriptor: | ANTI-IL-15 ANTIBODY, INTERLEUKIN 15, SULFATE ION | | Authors: | Lowe, D.C, Gerhardt, S, Ward, A, Hargreaves, D, Anderson, M, StGallay, S, Vousden, K, Ferraro, F, Pauptit, R.A, Cochrane, D, Pattison, D.V, Buchanan, C, Popovic, B, Finch, D.K, Wilkinson, T, Sleeman, M, Vaughan, T.J, Cruwys, S, Mallinder, P.R. | | Deposit date: | 2010-09-01 | | Release date: | 2010-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Engineering a High Affinity Anti-Il-15 Antibody: Crystal Structure Reveals an Alpha-Helix in Vh Cdr3 as Key Component of Paratope.
J.Mol.Biol., 406, 2011
|
|
5ITR
 
 | | Crystal Structure of Human NEIL1(P2G) bound to duplex DNA containing THF | | Descriptor: | DNA (26-MER), Endonuclease 8-like 1 | | Authors: | Zhu, C, Lu, L, Zhang, J, Yue, Z, Song, J, Zong, S, Liu, M, Stovicek, O, Gao, Y, Yi, C. | | Deposit date: | 2016-03-17 | | Release date: | 2016-07-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Tautomerization-dependent recognition and excision of oxidation damage in base-excision DNA repair
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8K8Z
 
 | | CryoEM structure of LonC protease hexamer in presence of AGS | | Descriptor: | Monothiophosphate, endopeptidase La | | Authors: | Li, M, Hsieh, K, Liu, H, Zhang, S, Gao, Y, Gong, Q, Zhang, K, Chang, C, Li, S. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis
Fundam Res, 2024
|
|
6Y1T
 
 | | The crystal structure of engineered cytochrome c peroxidase from Saccharomyces cerevisiae with a Trp51 to S-Trp51 modification | | Descriptor: | 1,2-ETHANEDIOL, Cytochrome c peroxidase, mitochondrial, ... | | Authors: | Ortmayer, M, Levy, C, Green, A.P. | | Deposit date: | 2020-02-13 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Noncanonical Tryptophan Analogue Reveals an Active Site Hydrogen Bond Controlling Ferryl Reactivity in a Heme Peroxidase.
Jacs Au, 1, 2021
|
|
2MUY
 
 | | The solution structure of the FtsH periplasmic N-domain | | Descriptor: | ATP-dependent zinc metalloprotease FtsH | | Authors: | Scharfenberg, F, Serek-Heuberger, J, Martin, J, Lupas, A.N, Coles, M. | | Deposit date: | 2014-09-18 | | Release date: | 2015-01-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and Evolution of N-domains in AAA Metalloproteases.
J.Mol.Biol., 427, 2015
|
|
6Y2Y
 
 | | The crystal structure of engineered cytochrome c peroxidase from Saccharomyces cerevisiae with Trp51 to S-Trp51 and Trp191Phe modifications | | Descriptor: | 1,2-ETHANEDIOL, Cytochrome c peroxidase, mitochondrial, ... | | Authors: | Ortmayer, M, Levy, C, Green, A.P. | | Deposit date: | 2020-02-17 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Noncanonical Tryptophan Analogue Reveals an Active Site Hydrogen Bond Controlling Ferryl Reactivity in a Heme Peroxidase.
Jacs Au, 1, 2021
|
|
5IY4
 
 | |
5H42
 
 | | Crystal Structure of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans in complex with alpha-d-glucose-1-phosphate | | Descriptor: | 1-O-phosphono-alpha-D-glucopyranose, Uncharacterized protein, alpha-D-glucopyranose | | Authors: | Nakajima, M, Tanaka, N, Furukawa, N, Nihira, T, Kodutsumi, Y, Takahashi, Y, Sugimoto, N, Miyanaga, A, Fushinobu, S, Taguchi, H, Nakai, H. | | Deposit date: | 2016-10-28 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanistic insight into the substrate specificity of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans
Sci Rep, 7, 2017
|
|
8K92
 
 | | CryoEM structure of LonC S582A hexamer with Lysozyme | | Descriptor: | Endopeptidase La, Monothiophosphate | | Authors: | Li, M, Hsieh, K, Liu, H, Zhang, S, Gao, Y, Gong, Q, Zhang, K, Chang, C, Li, S. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis
Fundam Res, 2024
|
|
2Y23
 
 | |
3VGG
 
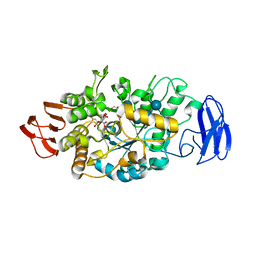 | | Crystal structure of glycosyltrehalose trehalohydrolase (E283Q) complexed with maltoheptaose | | Descriptor: | CITRATE ANION, GLYCEROL, Malto-oligosyltrehalose trehalohydrolase, ... | | Authors: | Okazaki, N, Tamada, T, Feese, M.D, Kato, M, Miura, Y, Komeda, T, Kobayashi, K, Kondo, K, Kuroki, R. | | Deposit date: | 2011-08-09 | | Release date: | 2012-06-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Substrate recognition mechanism of a glycosyltrehalose trehalohydrolase from Sulfolobus solfataricus KM1.
Protein Sci., 21, 2012
|
|
8K95
 
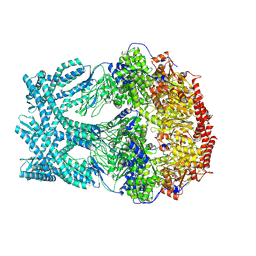 | | CryoEM structure of LonC protease open Hexamer, AGS | | Descriptor: | Monothiophosphate, endopeptidase La | | Authors: | Li, M, Hsieh, K, Liu, H, Zhang, S, Gao, Y, Gong, Q, Zhang, K, Chang, C, Li, S. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis
Fundam Res, 2024
|
|
8K91
 
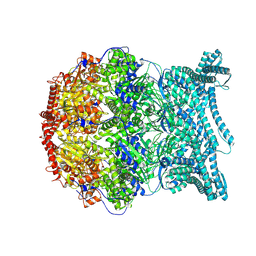 | | CryoEM structure of LonC S582A hepatmer with Lysozyme | | Descriptor: | Endopeptidase La, Monothiophosphate | | Authors: | Li, M, Hsieh, K, Liu, H, Zhang, S, Gao, Y, Gong, Q, Zhang, K, Chang, C, Li, S. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis
Fundam Res, 2024
|
|
6YR7
 
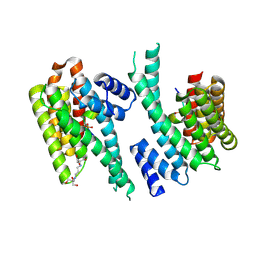 | | 14-3-3 sigma in complex with hDMX-342+367 peptide | | Descriptor: | 14-3-3 protein sigma, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Protein Mdm4 | | Authors: | Wolter, M, Srdanovic, S, Warriner, S, Wilson, A, Ottmann, C. | | Deposit date: | 2020-04-19 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | Understanding the interaction of 14-3-3 proteins with hDMX and hDM2: a structural and biophysical study.
Febs J., 2022
|
|
5IZN
 
 | | The crystal structure of 50S ribosomal protein L25 from Vibrio vulnificus CMCP6 | | Descriptor: | 50S ribosomal protein L25, PHOSPHATE ION | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-03-25 | | Release date: | 2016-04-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The crystal structure of 50S ribosomal protein L25 from Vibrio vulnificus CMCP6
To Be Published
|
|
6YR5
 
 | | 14-3-3 sigma in complex with hDMX-367 peptide | | Descriptor: | 14-3-3 protein sigma, Protein Mdm4, SULFATE ION | | Authors: | Wolter, M, Srdanovic, S, Ottman, C, Warriner, S, Wilson, A. | | Deposit date: | 2020-04-19 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Understanding the interaction of 14-3-3 proteins with hDMX and hDM2: a structural and biophysical study.
Febs J., 2022
|
|
6YR6
 
 | | 14-3-3 sigma in complex with hDM2-186 peptide | | Descriptor: | 14-3-3 protein sigma, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, IODIDE ION, ... | | Authors: | Wolter, M, Srdanovic, S, Warriner, S, Wilson, A, Ottmann, C. | | Deposit date: | 2020-04-19 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Understanding the interaction of 14-3-3 proteins with hDMX and hDM2: a structural and biophysical study.
Febs J., 2022
|
|
8K93
 
 | | CryoEM structure of LonC protease S582A open hexamer with lysozyme | | Descriptor: | Endopeptidase La, Monothiophosphate | | Authors: | Li, M, Hsieh, K, Liu, H, Zhang, S, Gao, Y, Gong, Q, Zhang, K, Chang, C, Li, S. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis
Fundam Res, 2024
|
|
8K8Y
 
 | | CryoEM structure of LonC heptamer in presence of AGS | | Descriptor: | Monothiophosphate, endopeptidase La | | Authors: | Li, M, Hsieh, K, Liu, H, Zhang, S, Gao, Y, Gong, Q, Zhang, K, Chang, C, Li, S. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis
Fundam Res, 2024
|
|
