3C5M
 
 | | Crystal structure of oligogalacturonate lyase (VPA0088) from Vibrio parahaemolyticus. Northeast Structural Genomics Consortium Target VpR199 | | Descriptor: | MANGANESE (II) ION, Oligogalacturonate lyase | | Authors: | Forouhar, F, Abashidze, M, Seetharaman, J, Janjua, H, Mao, L, Xiao, R, Owens, L.A, Wang, D, Baran, M.C, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-01-31 | | Release date: | 2008-02-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of oligogalacturonate lyase (VPA0088) from Vibrio parahaemolyticus.
To be Published
|
|
3BYP
 
 | | Mode of Action of a Putative Zinc Transporter CzrB | | Descriptor: | CzrB protein, SULFATE ION | | Authors: | Cherezov, V, Srinivasan, V, Szebenyi, D.M.E, Caffrey, M. | | Deposit date: | 2008-01-16 | | Release date: | 2008-09-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Insights into the Mode of Action of a Putative Zinc Transporter CzrB in Thermus thermophilus
Structure, 16, 2008
|
|
3BYV
 
 | | Crystal structure of Toxoplasma gondii specific rhoptry antigen kinase domain | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Rhoptry kinase | | Authors: | Wernimont, A.K, Lunin, V.V, Yang, C, Lew, J, Kozieradzki, I, Lin, Y.H, Sun, X, Khuu, C, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bochkarev, A, Hui, R, Sibley, D, Qiu, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-01-16 | | Release date: | 2008-01-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Novel structural and regulatory features of rhoptry secretory kinases in Toxoplasma gondii.
Embo J., 28, 2009
|
|
1QK1
 
 | | CRYSTAL STRUCTURE OF HUMAN UBIQUITOUS MITOCHONDRIAL CREATINE KINASE | | Descriptor: | CREATINE KINASE, UBIQUITOUS MITOCHONDRIAL, PHOSPHATE ION | | Authors: | Eder, M, Schlattner, U, Fritz-Wolf, K, Wallimann, T, Kabsch, W. | | Deposit date: | 1999-07-08 | | Release date: | 2000-04-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Human Ubiquitous Mitochondrial Creatine Kinase
Proteins: Struct.,Funct., Genet., 39, 2000
|
|
2QF8
 
 | | Crystal structure of the complex of Buffalo Secretory Glycoprotein with tetrasaccharide at 2.8A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase-3-like protein 1 | | Authors: | Singh, A.K, Jain, R, Sinha, M, Kumar, A, Singh, N, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2007-06-27 | | Release date: | 2007-07-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the complex of Buffalo Secretory Glycoprotein with Tetrasaccharide at 2.8A resolution
To be Published
|
|
2QFL
 
 | | Structure of SuhB: Inositol monophosphatase and extragenic suppressor from E. coli | | Descriptor: | ACETATE ION, ETHYL ACETATE, Inositol-1-monophosphatase | | Authors: | Wang, Y, Stieglitz, K.A, Bubunenko, M, Court, D, Stec, B, Roberts, M.F. | | Deposit date: | 2007-06-27 | | Release date: | 2007-07-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of the R184A mutant of the inositol monophosphatase encoded by suhB and implications for its functional interactions in Escherichia coli.
J.Biol.Chem., 282, 2007
|
|
3C0B
 
 | | Crystal structure of the conserved archaeal protein Q6M145. Northeast Structural Genomics Consortium target MrR63 | | Descriptor: | CALCIUM ION, Conserved archaeal protein Q6M145 | | Authors: | Kuzin, A.P, Su, M, Seetharaman, J, Wang, D, Fang, Y, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-01-19 | | Release date: | 2008-02-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray structure of the conserved archaeal protein Q6M145.
To be Published
|
|
2QCO
 
 | | Crystal structure of the transcriptional regulator CmeR from Campylobacter jejuni | | Descriptor: | CmeR, GLYCEROL | | Authors: | Gu, R, Su, C, Shi, F, Li, M, McDermott, G, Zhang, Q, Yu, E.W. | | Deposit date: | 2007-06-19 | | Release date: | 2008-04-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the transcriptional regulator CmeR from Campylobacter jejuni.
J.Mol.Biol., 372, 2007
|
|
1HLO
 
 | | THE CRYSTAL STRUCTURE OF AN INTACT HUMAN MAX-DNA COMPLEX: NEW INSIGHTS INTO MECHANISMS OF TRANSCRIPTIONAL CONTROL | | Descriptor: | DNA (5'-D(*AP*CP*CP*AP*CP*GP*TP*GP*GP*TP*G)-3'), DNA (5'-D(*CP*AP*CP*CP*AP*CP*GP*TP*GP*GP*T)-3'), PROTEIN (TRANSCRIPTION FACTOR MAX) | | Authors: | Brownlie, P, Ceska, T.A, Lamers, M, Romier, C, Theo, H, Suck, D. | | Deposit date: | 1997-09-10 | | Release date: | 1997-10-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of an intact human Max-DNA complex: new insights into mechanisms of transcriptional control.
Structure, 5, 1997
|
|
1HQP
 
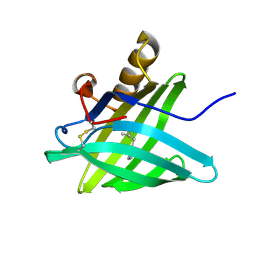 | | CRYSTAL STRUCTURE OF A TRUNCATED FORM OF PORCINE ODORANT-BINDING PROTEIN | | Descriptor: | 2-ISOBUTYL-3-METHOXYPYRAZINE, ODORANT-BINDING PROTEIN | | Authors: | Perduca, M, Mancia, F, Del Giorgio, R, Monaco, H.L. | | Deposit date: | 2000-12-19 | | Release date: | 2001-01-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a truncated form of porcine odorant-binding protein.
Proteins, 42, 2001
|
|
1HN2
 
 | | CRYSTAL STRUCTURE OF BOVINE OBP COMPLEXED WITH AMINOANTHRACENE | | Descriptor: | (3R)-oct-1-en-3-ol, ANTHRACEN-1-YLAMINE, ODORANT-BINDING PROTEIN | | Authors: | Vincent, F, Spinelli, S, Tegoni, M, Cambillau, C. | | Deposit date: | 2000-12-05 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The insect attractant 1-octen-3-ol is the natural ligand of bovine odorant-binding protein.
J.Biol.Chem., 276, 2001
|
|
1QJS
 
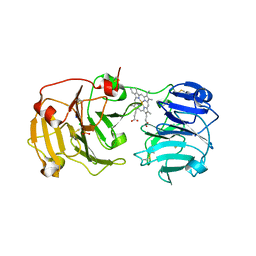 | | mammalian blood serum haemopexin glycosylated-native protein and in complex with its ligand haem | | Descriptor: | CHLORIDE ION, HEMOPEXIN, PHOSPHATE ION, ... | | Authors: | Paoli, M, Baker, H.M, Morgan, W.T, Smith, A, Baker, E.N. | | Deposit date: | 1999-07-01 | | Release date: | 2000-02-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Hemopexin Reveals a Novel High-Affinity Heme Site Formed between Two Beta-Propeller Domains.
Nat.Struct.Biol., 6, 1999
|
|
2QGA
 
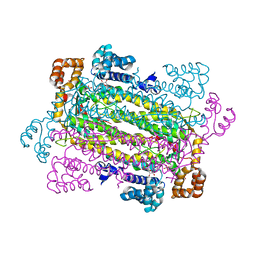 | | Plasmodium vivax adenylosuccinate lyase Pv003765 with AMP bound | | Descriptor: | ADENOSINE MONOPHOSPHATE, Adenylosuccinate lyase, CALCIUM ION, ... | | Authors: | Lunin, V.V, Wernimont, A.K, Lew, J, Kozieradzki, I, Bochkarev, A, Arrowsmith, C.H, Sundstrom, M, Weigelt, J, Edwards, A.E, Hui, R, Hills, T, Altamentova, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-28 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Plasmodium vivax adenylosuccinate lyase Pv003765 with AMP bound
To be Published
|
|
2QH7
 
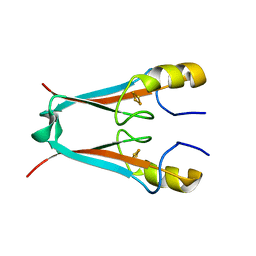 | | MitoNEET is a uniquely folded 2Fe-2S outer mitochondrial membrane protein stabilized by pioglitazone | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Zinc finger CDGSH-type domain 1 | | Authors: | Paddock, M.L, Wiley, S.E, Axelrod, H.L, Cohen, A.E, Roy, M, Abresch, E.C, Capraro, D, Murphy, A.N, Nechushtai, R, Dixon, J.E, Jennings, P.A. | | Deposit date: | 2007-06-30 | | Release date: | 2007-08-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | MitoNEET is a uniquely folded 2Fe 2S outer mitochondrial membrane protein stabilized by pioglitazone.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3ZKB
 
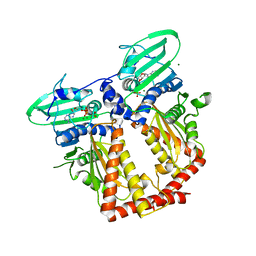 | | CRYSTAL STRUCTURE OF THE ATPASE REGION OF Mycobacterium tuberculosis GyrB WITH AMPPNP | | Descriptor: | DNA GYRASE SUBUNIT B, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Agrawal, A, Roue, M, Spitzfaden, C, Petrella, S, Aubry, A, Volker, C, Mossakowska, D, Hann, M, Bax, B, Mayer, C. | | Deposit date: | 2013-01-22 | | Release date: | 2013-09-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mycobacterium Tuberculosis DNA Gyrase ATPase Domain Structures Suggest a Dissociative Mechanism that Explains How ATP Hydrolysis is Coupled to Domain Motion.
Biochem.J., 456, 2013
|
|
1QO7
 
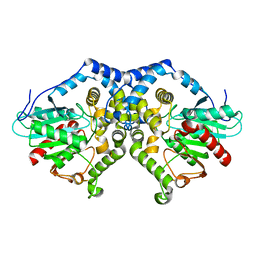 | | Structure of Aspergillus niger epoxide hydrolase | | Descriptor: | EPOXIDE HYDROLASE | | Authors: | Zou, J.-Y, Hallberg, B.M, Bergfors, T, Oesch, F, Arand, M, Mowbray, S.L, Jones, T.A. | | Deposit date: | 1999-11-04 | | Release date: | 2000-02-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Aspergillus Niger Epoxide Hydrolase at 1.8A Resolution: Implications for the Structure and Function of the Mammalian Microsomal Class of Epoxide Hydrolases
Structure, 8, 2000
|
|
2QKT
 
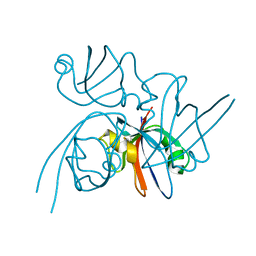 | |
2QKR
 
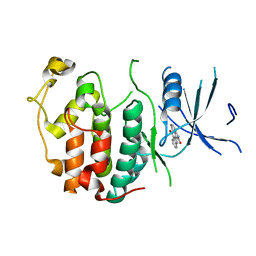 | | Cryptosporidium parvum cyclin-dependent kinase cgd5_2510 with indirubin 3'-monoxime bound | | Descriptor: | (Z)-1H,1'H-[2,3']BIINDOLYLIDENE-3,2'-DIONE-3-OXIME, Cdc2-like CDK2/CDC28 like protein kinase | | Authors: | Wernimont, A.K, Dong, A, Lew, J, Lin, Y.H, Hassanali, A, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Artz, J.D, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-07-11 | | Release date: | 2007-09-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Cryptosporidium parvum cyclin-dependent kinase cgd5_2510 with indirubin 3'-monoxime bound.
To be Published
|
|
3BWL
 
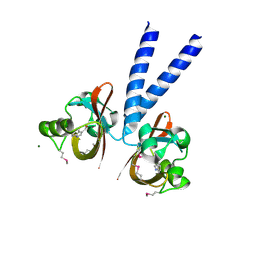 | | Crystal structure of PAS domain of HTR-like protein from Haloarcula marismortui | | Descriptor: | 1H-INDOLE-3-CARBALDEHYDE, MAGNESIUM ION, Sensor protein | | Authors: | Osipiuk, J, Zhou, M, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-01-09 | | Release date: | 2008-01-15 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | X-ray crystal structure of PAS domain of HTR-like protein from Haloarcula marismortui.
To be Published
|
|
1R5H
 
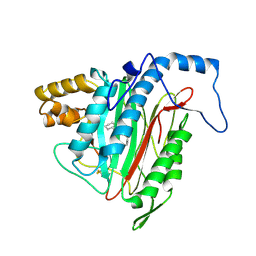 | | Crystal Structure of MetAP2 complexed with A320282 | | Descriptor: | MANGANESE (II) ION, Methionine aminopeptidase 2, N'-(2S,3R)-3-AMINO-4-CYCLOHEXYL-2-HYDROXY-BUTANO-N-(4-METHYLPHENYL)HYDRAZIDE | | Authors: | Sheppard, G.S, Wang, J, Kawai, M, BaMaung, N.Y, Craig, R.A, Erickson, S.A, Lynch, L, Patel, J, Yang, F, Searle, X.B, Lou, P, Park, C, Kim, K.H, Henkin, J, Lesniewski, R. | | Deposit date: | 2003-10-10 | | Release date: | 2004-10-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 3-Amino-2-hydroxyamides and related compounds as inhibitors of methionine aminopeptidase-2.
Bioorg.Med.Chem.Lett., 14, 2004
|
|
1R5S
 
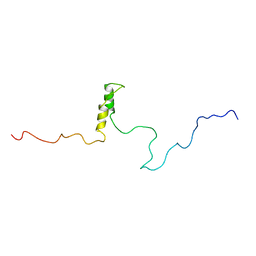 | | Connexin 43 Carboxyl Terminal Domain | | Descriptor: | Gap junction alpha-1 protein | | Authors: | Sorgen, P.L, Duffy, H.S, Mario, D, Sahoo, P, Coombs, W, Delmar, M, Spray, D.C. | | Deposit date: | 2003-10-13 | | Release date: | 2004-10-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural changes in the carboxyl terminus of the gap junction protein connexin43 indicates signaling between binding domains for c-Src and zonula occludens-1
J.Biol.Chem., 279, 2004
|
|
2QMW
 
 | | The crystal structure of the prephenate dehydratase (PDT) from Staphylococcus aureus subsp. aureus Mu50 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tan, K, Zhang, R, Li, H, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-17 | | Release date: | 2007-08-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of open (R) and close (T) states of prephenate dehydratase (PDT) - implication of allosteric regulation by L-phenylalanine.
J.Struct.Biol., 162, 2008
|
|
2QR8
 
 | |
3BZO
 
 | | Crystal structural of native EscU C-terminal domain | | Descriptor: | EscU, SULFATE ION | | Authors: | Zarivach, R, Deng, W, Vuckovic, M, Felise, H.B, Nguyen, H.V, Miller, S.I, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2008-01-18 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural analysis of the essential self-cleaving type III secretion proteins EscU and SpaS.
Nature, 453, 2008
|
|
3BZV
 
 | | Crystal structural of the mutated T264A EscU C-terminal domain | | Descriptor: | EscU | | Authors: | Zarivach, R, Deng, W, Vuckovic, M, Felise, H.B, Nguyen, H.V, Miller, S.I, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2008-01-18 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural analysis of the essential self-cleaving type III secretion proteins EscU and SpaS.
Nature, 453, 2008
|
|
