7CD1
 
 | | Crystal structure of inhibitory Smad, Smad7 | | Descriptor: | CHLORIDE ION, Mothers against decapentaplegic homolog 7, SULFATE ION | | Authors: | Murayama, K, Kato-Murayama, M, Shirouzu, M. | | Deposit date: | 2020-06-18 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural basis for inhibitory effects of Smad7 on TGF-beta family signaling.
J.Struct.Biol., 212, 2020
|
|
3MML
 
 | | Allophanate Hydrolase Complex from Mycobacterium smegmatis, Msmeg0435-Msmeg0436 | | Descriptor: | Allophanate hydrolase subunit 1, Allophanate hydrolase subunit 2, CHLORIDE ION | | Authors: | Kaufmann, M, Chernishof, I, Shin, A, Germano, D, Sawaya, M.R, Waldo, G.S, Arbing, M.A, Perry, J, Eisenberg, D, Integrated Center for Structure and Function Innovation (ISFI), TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-04-20 | | Release date: | 2010-04-28 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Allphanate Hydrolase Complex from M. smegmatis, Msmeg0435-Msmeg0436
To be Published
|
|
1K5N
 
 | | HLA-B*2709 BOUND TO NONA-PEPTIDE M9 | | Descriptor: | GLYCEROL, beta-2-microglobulin, light chain, ... | | Authors: | Hulsmeyer, M, Hillig, R.C, Volz, A, Ruhl, M, Schroder, W, Saenger, W, Ziegler, A, Uchanska-Ziegler, B. | | Deposit date: | 2001-10-11 | | Release date: | 2002-10-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | HLA-B27 Subtypes Differentially Associated with Disease Exhibit Subtle Structural Alterations
J.Biol.Chem., 277, 2002
|
|
7MDH
 
 | | STRUCTURAL BASIS FOR LIGHT ACITVATION OF A CHLOROPLAST ENZYME. THE STRUCTURE OF SORGHUM NADP-MALATE DEHYDROGENASE IN ITS OXIDIZED FORM | | Descriptor: | PROTEIN (MALATE DEHYDROGENASE), ZINC ION | | Authors: | Johansson, K, Ramaswamy, S, Saarinen, M, Lemaire-Chamley, M, Issakidis-Bourguet, E, Miginiac-Maslow, M, Eklund, H. | | Deposit date: | 1999-02-16 | | Release date: | 1999-06-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for light activation of a chloroplast enzyme: the structure of sorghum NADP-malate dehydrogenase in its oxidized form.
Biochemistry, 38, 1999
|
|
4LB3
 
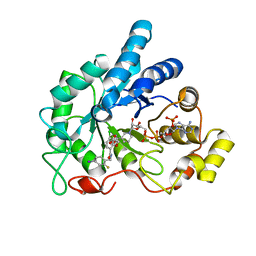 | | Crystal structure of human AR complexed with NADP+ and {5-chloro-2-[(2-fluoro-4-iodobenzyl)carbamoyl]phenoxy}acetic acid | | Descriptor: | Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, {5-chloro-2-[(2-fluoro-4-iodobenzyl)carbamoyl]phenoxy}acetic acid | | Authors: | Cousido-Siah, A, Mitschler, A, Ruiz, F.X, Fanfrlik, J, Kolar, M, Hobza, P, Podjarny, A. | | Deposit date: | 2013-06-20 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Modulation of aldose reductase inhibition by halogen bond tuning.
Acs Chem.Biol., 8, 2013
|
|
3MRE
 
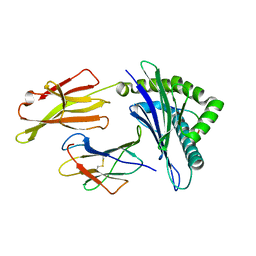 | | Crystal Structure of MHC class I HLA-A2 molecule complexed with EBV bmlf1-280-288 nonapeptide | | Descriptor: | 9-meric peptide from mRNA export factor EB2, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Gras, S, Echasserieau, K, Saulquin, X, Bonneville, M, Housset, D. | | Deposit date: | 2010-04-29 | | Release date: | 2011-05-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Analysis of Relationships between Peptide/MHC Structural Features and Naive T Cell Frequency in Humans.
J.Immunol., 193, 2014
|
|
3MRL
 
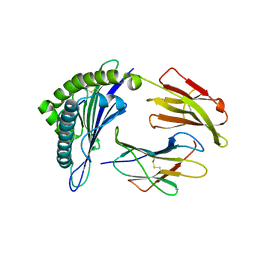 | | Crystal Structure of MHC class I HLA-A2 molecule complexed with HCV NS3-1073-1081 nonapeptide C6V variant | | Descriptor: | 9-meric peptide from Serine protease/NTPase/helicase NS3, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Gras, S, Reiser, J.-B, Chouquet, A, Le Gorrec, M, Debeaupuis, E, Echasserieau, K, Saulquin, X, Bonneville, M, Housset, D. | | Deposit date: | 2010-04-29 | | Release date: | 2011-05-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Analysis of Relationships between Peptide/MHC Structural Features and Naive T Cell Frequency in Humans.
J.Immunol., 193, 2014
|
|
4PVV
 
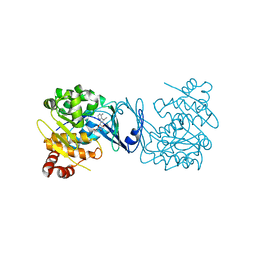 | | Micobacterial Adenosine Kinase in complex with inhibitor | | Descriptor: | 5-ethynyl-7-(beta-D-ribofuranosyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Adenosine kinase | | Authors: | Pichova, I, Hocek, M, Dostal, J, Rezacova, P. | | Deposit date: | 2014-03-18 | | Release date: | 2014-11-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Inhibition of Mycobacterial and Human Adenosine Kinase by 7-Substituted 7-(Het)aryl-7-deazaadenine Ribonucleosides
J.Med.Chem., 57, 2014
|
|
3NDA
 
 | | Crystal structure of serpin from tick Ixodes ricinus | | Descriptor: | PENTAETHYLENE GLYCOL, Serpin-2 | | Authors: | Rezacova, P, Kovarova, Z, Chmelar, J, Mares, M. | | Deposit date: | 2010-06-07 | | Release date: | 2010-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A tick salivary protein targets cathepsin G and chymase and inhibits host inflammation and platelet aggregation.
Blood, 117, 2011
|
|
7CTN
 
 | | Structure of the 328-692 fragment of FlhA (E351A/D356A) | | Descriptor: | Flagellar biosynthesis protein FlhA | | Authors: | Kida, M, Takekawa, N, Kinoshita, M, Inoue, Y, Minamino, T, Imada, K. | | Deposit date: | 2020-08-19 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The FlhA linker mediates flagellar protein export switching during flagellar assembly.
Commun Biol, 4, 2021
|
|
3NHK
 
 | |
3VT4
 
 | | Crystal structures of rat VDR-LBD with R270L mutation | | Descriptor: | (1R,2Z,3R,5E,7E)-17-{(1S)-1-[(2-ethyl-2-hydroxybutyl)sulfanyl]ethyl}-2-(2-hydroxyethylidene)-9,10-secoestra-5,7,16-triene-1,3-diol, COACTIVATOR PEPTIDE DRIP, Vitamin D3 receptor | | Authors: | Nakabayashi, M, Shimizu, M, Ikura, T, Ito, N. | | Deposit date: | 2012-05-19 | | Release date: | 2013-05-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of hereditary vitamin D-resistant rickets-associated vitamin D receptor mutants R270L and W282R bound to 1,25-dihydroxyvitamin D3 and synthetic ligands.
J.Med.Chem., 56, 2013
|
|
7CG3
 
 | | Staggered ring conformation of CtHsp104 (Hsp104 from Chaetomium Thermophilum) | | Descriptor: | Heat shock protein 104 | | Authors: | Inoue, Y, Hanazono, Y, Noi, K, Kawamoto, A, Kimatsuka, M, Harada, R, Takeda, K, Iwamasa, N, Shibata, K, Noguchi, K, Shigeta, Y, Namba, K, Ogura, T, Miki, K, Shinohara, K, Yohda, M. | | Deposit date: | 2020-06-30 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Split conformation of Chaetomium thermophilum Hsp104 disaggregase.
Structure, 29, 2021
|
|
3NBT
 
 | | Crystal structure of trimeric cytochrome c from horse heart | | Descriptor: | Cytochrome c, DI(HYDROXYETHYL)ETHER, HEME C, ... | | Authors: | Taketa, M, Komori, H, Hirota, S, Higuchi, Y. | | Deposit date: | 2010-06-04 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cytochrome c polymerization by successive domain swapping at the C-terminal helix
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4LB4
 
 | | Crystal structure of human AR complexed with NADP+ and {2-[(4-bromo-2,3,5,6-tetrafluorobenzyl)carbamoyl]-5-chlorophenoxy}acetic acid | | Descriptor: | Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, {2-[(4-bromo-2,3,5,6-tetrafluorobenzyl)carbamoyl]-5-chlorophenoxy}acetic acid | | Authors: | Cousido-Siah, A, Mitschler, A, Ruiz, F.X, Fanfrlik, J, Kolar, M, Hobza, P, Podjarny, A. | | Deposit date: | 2013-06-20 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Modulation of aldose reductase inhibition by halogen bond tuning.
Acs Chem.Biol., 8, 2013
|
|
3NI7
 
 | | Crystal structure of the TetR transcriptional regulator from Nitrosomonas europaea ATCC 19718 | | Descriptor: | Bacterial regulatory proteins, TetR family | | Authors: | Knapik, A, Chruszcz, M, Cymborowski, M, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-15 | | Release date: | 2010-08-11 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystal structure of the TetR transcriptional regulator from Nitrosomonas europaea ATCC 19718
To be Published
|
|
2Z3J
 
 | | Crystal structure of blasticidin S deaminase (BSD) R90K mutant | | Descriptor: | Blasticidin-S deaminase, CACODYLATE ION, CHLORIDE ION, ... | | Authors: | Teh, A.H, Kumasaka, T, Yamamoto, M, Furuichi, M, Nakasako, M, Kimura, M, Yamaguchi, I, Ueki, T. | | Deposit date: | 2007-06-04 | | Release date: | 2007-10-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of blasticidin S deaminase (BSD): implications for dynamic properties of catalytic zinc
J.Biol.Chem., 282, 2007
|
|
3N6V
 
 | |
4K91
 
 | | Crystal structure of Penicillin-Binding Protein 5 (PBP5) from Pseudomonas aeruginosa in apo state | | Descriptor: | D-ala-D-ala-carboxypeptidase, SUCCINIC ACID | | Authors: | Smith, J, Toth, M, Vakulenko, S, Mobashery, S, Chen, Y. | | Deposit date: | 2013-04-19 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural analysis of the role of Pseudomonas aeruginosa penicillin-binding protein 5 in beta-lactam resistance.
Antimicrob.Agents Chemother., 57, 2013
|
|
3ETF
 
 | | Crystal structure of a putative succinate-semialdehyde dehydrogenase from salmonella typhimurium lt2 | | Descriptor: | Putative succinate-semialdehyde dehydrogenase | | Authors: | Brunzelle, J.S, Evdokimova, E, Kudritska, M, Wawrzak, Z, Anderson, W.F, Savchenk, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-10-07 | | Release date: | 2008-11-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and activity of the NAD(P)(+) -dependent succinate semialdehyde dehydrogenase YneI from Salmonella typhimurium.
Proteins, 81, 2013
|
|
3VWU
 
 | |
3MRD
 
 | | Crystal Structure of MHC class I HLA-A2 molecule complexed with HCMV pp65-495-503 nonapeptide V6G variant | | Descriptor: | 9-meric peptide from Tegument protein pp65, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Gras, S, Reiser, J.-B, Chouquet, A, Debeaupuis, E, Echasserieau, K, Saulquin, X, Bonneville, M, Housset, D. | | Deposit date: | 2010-04-29 | | Release date: | 2011-05-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Analysis of Relationships between Peptide/MHC Structural Features and Naive T Cell Frequency in Humans.
J.Immunol., 193, 2014
|
|
3MRJ
 
 | | Crystal Structure of MHC class I HLA-A2 molecule complexed with HCV NS3-1073-1081 nonapeptide V5M variant | | Descriptor: | 9-meric peptide from Serine protease/NTPase/helicase NS3, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Reiser, J.-B, Le Gorrec, M, Chouquet, A, Debeaupuis, E, Echasserieau, K, Saulquin, X, Bonneville, M, Housset, D. | | Deposit date: | 2010-04-29 | | Release date: | 2011-05-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal Structure of MHC class I HLA-A2 molecule complexed with HCV NS3-1073-1081 nonapeptide V5M variant
To be Published
|
|
3E9V
 
 | | Crystal structure of human B-cell Translocation Gene 2 (BTG2) | | Descriptor: | 1,2-ETHANEDIOL, Protein BTG2 | | Authors: | Sampathkumar, P, Romero, R, Wasserman, S, Hu, S, Maletic, M, Freeman, J, Tarun, G, Atwell, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-08-23 | | Release date: | 2008-10-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of human B-cell Translocation Gene 2 (BTG2)
To be Published
|
|
1K7V
 
 | | Crystal Structure Analysis of crosslinked-WGA3/GlcNAcbeta1,6Galbeta1,4Glc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, agglutinin isolectin 3 | | Authors: | Muraki, M, Ishimura, M, Harata, K. | | Deposit date: | 2001-10-22 | | Release date: | 2002-04-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Interactions of wheat-germ agglutinin with GlcNAc beta 1,6Gal sequence
Biochim.Biophys.Acta, 1569, 2002
|
|
