6SRL
 
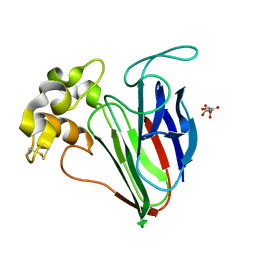 | |
8FTQ
 
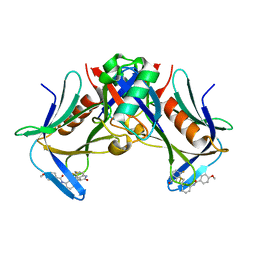 | | Crystal structure of hRpn13 Pru domain in complex with Ubiquitin and XL44 | | Descriptor: | N-(3-{[(3R)-5-fluoro-2-oxo-2,3-dihydro-1H-indol-3-yl]methyl}phenyl)-4-methoxybenzamide, Proteasomal ubiquitin receptor ADRM1, Ubiquitin | | Authors: | Walters, K.J, Lu, X, Chandravanshi, M. | | Deposit date: | 2023-01-13 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A structure-based designed small molecule depletes hRpn13 Pru and a select group of KEN box proteins.
Nat Commun, 15, 2024
|
|
6SRT
 
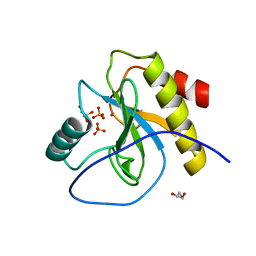 | | Endolysine N-acetylmuramoyl-L-alanine amidase LysCS from Clostridium intestinale URNW | | Descriptor: | GLYCEROL, N-acetylmuramoyl-L-alanine amidase, PHOSPHATE ION, ... | | Authors: | Hakansson, M, Al-Karadaghi, S, Plotka, M, Kaczorowska, A.-K, Kaczorowski, T. | | Deposit date: | 2019-09-06 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Structure and function of endolysines LysCS, LysC from Clostridium intestinale
To Be Published
|
|
5H65
 
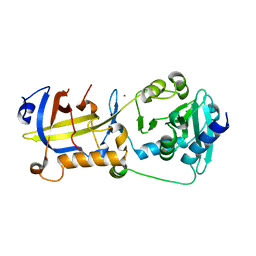 | | Crystal structure of human POT1 and TPP1 | | Descriptor: | Adrenocortical dysplasia protein homolog, Protection of telomeres protein 1, ZINC ION | | Authors: | Chen, C, Wu, J, Lei, M. | | Deposit date: | 2016-11-10 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into POT1-TPP1 interaction and POT1 C-terminal mutations in human cancer.
Nat Commun, 8, 2017
|
|
1I4J
 
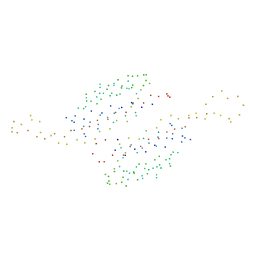 | | CRYSTAL STRUCTURE OF L22 RIBOSOMAL PROTEIN MUTANT | | Descriptor: | 50S RIBOSOMAL PROTEIN L22 | | Authors: | Davydova, N.L, Streltsov, V.A, Fedorov, R, Wilce, M, Liljas, A, Garder, M. | | Deposit date: | 2001-02-22 | | Release date: | 2002-09-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | L22 ribosomal protein and effect of its mutation on ribosome resistance to erythromycin.
J.Mol.Biol., 322, 2002
|
|
6GM5
 
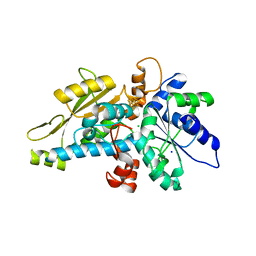 | | [FeFe]-hydrogenase HydA1 from Chlamydomonas reinhardtii,variant E141A | | Descriptor: | CHLORIDE ION, Fe-hydrogenase, IRON/SULFUR CLUSTER, ... | | Authors: | Duan, J, Engelbrecht, V, Esselborn, J, Hofmann, E, Winkler, M, Happe, T. | | Deposit date: | 2018-05-24 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystallographic and spectroscopic assignment of the proton transfer pathway in [FeFe]-hydrogenases.
Nat Commun, 9, 2018
|
|
6GO7
 
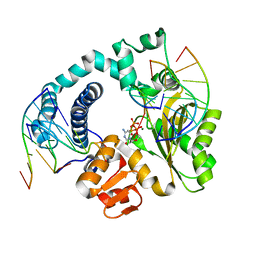 | | TdT chimera (Loop1 of pol mu) - full DNA synapsis complex | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*AP*AP*AP*C)-3'), DNA (5'-D(*TP*TP*TP*TP*TP*GP*C)-3'), ... | | Authors: | Loc'h, J, Gerodimos, C.A, Rosario, S, Lieber, M.R, Delarue, M. | | Deposit date: | 2018-06-01 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural evidence for an intransbase selection mechanism involving Loop1 in polymerase mu at an NHEJ double-strand break junction.
J.Biol.Chem., 294, 2019
|
|
6HXA
 
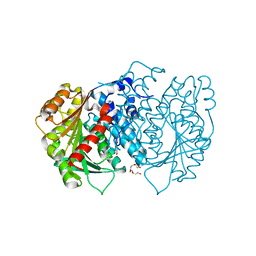 | | AntI from P. luminescens catalyses terminal polyketide shortening in the biosynthesis of anthraquinones | | Descriptor: | AntI, TETRAETHYLENE GLYCOL, TRIETHYLENE GLYCOL | | Authors: | Zhou, Q, Braeuer, A, Grammbitter, G, Schmalhofer, M, Saura, P, Adihou, H, Kaila, V.R.I, Groll, M, Bode, H. | | Deposit date: | 2018-10-16 | | Release date: | 2019-08-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular mechanism of polyketide shortening in anthraquinone biosynthesis ofPhotorhabdus luminescens.
Chem Sci, 10, 2019
|
|
6HY3
 
 | | Three-dimensional structure of AgaC from Zobellia galactanivorans | | Descriptor: | 1,2-ETHANEDIOL, Beta-agarase C, GLYCEROL, ... | | Authors: | Naretto, A, Fanuel, M, Ropartz, D, Rogniaux, H, Larocque, R, Czjzek, M, Tellier, C, Michel, G. | | Deposit date: | 2018-10-19 | | Release date: | 2019-03-13 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The agar-specific hydrolaseZgAgaC from the marine bacteriumZobellia galactanivoransdefines a new GH16 protein subfamily.
J.Biol.Chem., 294, 2019
|
|
6H3E
 
 | | Receptor-bound Ghrelin conformation | | Descriptor: | Appetite-regulating hormone, octan-1-amine | | Authors: | Ferre, G, Damian, M, M'Kadmi, C, Saurel, O, Czaplicki, G, Demange, P, Marie, J, Fehrentz, J.A, Baneres, J.L, Milon, A. | | Deposit date: | 2018-07-18 | | Release date: | 2019-07-24 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of G protein-coupled receptor-bound ghrelin reveal the critical role of the octanoyl chain.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7A7D
 
 | | Cadherin fit into cryo-ET map | | Descriptor: | Desmocollin-2, Desmoglein-2 | | Authors: | Sikora, M, Ermel, U.H, Seybold, A, Kunz, M, Calloni, G, Reitz, J, Vabulas, R.M, Hummer, G, Frangakis, A.S. | | Deposit date: | 2020-08-28 | | Release date: | 2020-12-23 | | Method: | ELECTRON MICROSCOPY (26 Å) | | Cite: | Desmosome architecture derived from molecular dynamics simulations and cryo-electron tomography.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2VGM
 
 | |
6SZA
 
 | |
2VGN
 
 | | Structure of S. cerevisiae Dom34, a translation termination-like factor involved in RNA quality control pathways and interacting with Hbs1 (SelenoMet-labeled protein) | | Descriptor: | DOM34, GLYCEROL, PHOSPHATE ION | | Authors: | Graille, M, Chaillet, M, Van Tilbeurgh, H. | | Deposit date: | 2007-11-14 | | Release date: | 2008-01-22 | | Last modified: | 2021-03-17 | | Method: | X-RAY DIFFRACTION (2.505 Å) | | Cite: | Structure of Yeast Dom34: A Protein Related to Translation Termination Factor Erf1 and Involved in No-Go Decay.
J.Biol.Chem., 283, 2008
|
|
2VGO
 
 | | Crystal structure of Aurora B kinase in complex with Reversine inhibitor | | Descriptor: | INNER CENTROMERE PROTEIN A, N~6~-cyclohexyl-N~2~-(4-morpholin-4-ylphenyl)-9H-purine-2,6-diamine, SERINE/THREONINE-PROTEIN KINASE 12-A | | Authors: | D'Alise, A.M, Amabile, G, Iovino, M, Di Giorgio, F.P, Bartiromo, M, Sessa, F, Villa, F, Musacchio, A, Cortese, R. | | Deposit date: | 2007-11-15 | | Release date: | 2008-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Reversine, a Novel Aurora Kinases Inhibitor, Inhibits Colony Formation of Human Acute Myeloid Leukemia Cells.
Mol.Cancer Ther., 7, 2008
|
|
1I96
 
 | | CRYSTAL STRUCTURE OF THE 30S RIBOSOMAL SUBUNIT FROM THERMUS THERMOPHILUS IN COMPLEX WITH THE TRANSLATION INITIATION FACTOR IF3 (C-TERMINAL DOMAIN) | | Descriptor: | 16S RRNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Pioletti, M, Schluenzen, F, Harms, J, Zarivach, R, Gluehmann, M, Avila, H, Bartels, H, Jacobi, C, Hartsch, T, Yonath, A, Franceschi, F. | | Deposit date: | 2001-03-18 | | Release date: | 2001-04-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Crystal structures of complexes of the small ribosomal subunit with tetracycline, edeine and IF3.
EMBO J., 20, 2001
|
|
5NPA
 
 | | Solution structure of Drosophila melanogaster Loquacious dsRBD2 | | Descriptor: | Loquacious | | Authors: | Tants, J.-N, Fesser, S, Kern, T, Stehle, R, Geerlof, A, Wunderlich, C, Boettcher, R, Kunzelmann, S, Lange, O, Kreutz, C, Foerstemann, K, Sattler, M. | | Deposit date: | 2017-04-16 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for asymmetry sensing of siRNAs by the Drosophila Loqs-PD/Dcr-2 complex in RNA interference.
Nucleic Acids Res., 45, 2017
|
|
6HYJ
 
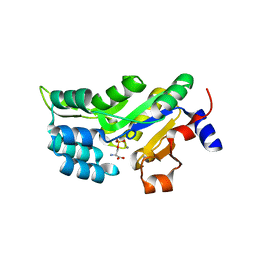 | | PSPH Human phosphoserine phosphatase | | Descriptor: | CALCIUM ION, PHOSPHOSERINE, Phosphoserine phosphatase, ... | | Authors: | Wouters, J, Haufroid, M, Mirgaux, M. | | Deposit date: | 2018-10-22 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.929 Å) | | Cite: | Crystal structures and snapshots along the reaction pathway of human phosphoserine phosphatase.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
5NPG
 
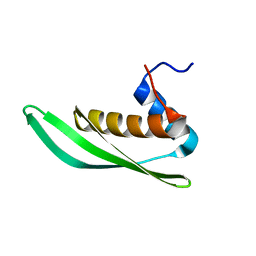 | | Solution structure of Drosophila melanogaster Loquacious dsRBD1 | | Descriptor: | Loquacious, isoform F | | Authors: | Tants, J.-N, Fesser, S, Kern, T, Stehle, R, Geerlof, A, Wunderlich, C, Hartlmuller, C, Boettcher, R, Kunzelmann, S, Lange, O, Kreutz, C, Foerstemann, K, Sattler, M. | | Deposit date: | 2017-04-16 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for asymmetry sensing of siRNAs by the Drosophila Loqs-PD/Dcr-2 complex in RNA interference.
Nucleic Acids Res., 45, 2017
|
|
5JKM
 
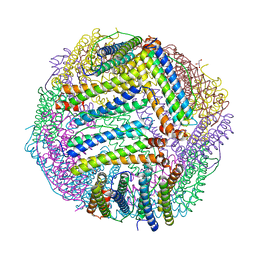 | |
6SKO
 
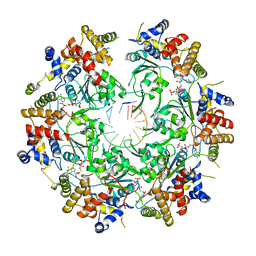 | | Cryo-EM Structure of the Fork Protection Complex Bound to CMG at a Replication Fork - conformation 2 MCM CTD:ssDNA | | Descriptor: | DNA replication licensing factor MCM2, DNA replication licensing factor MCM3, DNA replication licensing factor MCM4, ... | | Authors: | Yeeles, J, Baretic, D, Jenkyn-Bedford, M. | | Deposit date: | 2019-08-16 | | Release date: | 2020-05-06 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM Structure of the Fork Protection Complex Bound to CMG at a Replication Fork.
Mol.Cell, 78, 2020
|
|
6GM6
 
 | | [FeFe]-hydrogenase HydA1 from Chlamydomonas reinhardtii,variant E141Q | | Descriptor: | CHLORIDE ION, Fe-hydrogenase, IRON/SULFUR CLUSTER, ... | | Authors: | Duan, J, Engelbrecht, V, Esselborn, J, Hofmann, E, Winkler, M, Happe, T. | | Deposit date: | 2018-05-24 | | Release date: | 2018-11-07 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystallographic and spectroscopic assignment of the proton transfer pathway in [FeFe]-hydrogenases.
Nat Commun, 9, 2018
|
|
8HAP
 
 | | Crystal structure of thermostable acetaldehyde dehydrogenase from hyperthermophilic archaeon Sulfolobus tokodaii | | Descriptor: | 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, Aldehyde dehydrogenase, SODIUM ION, ... | | Authors: | Mine, S, Nakabayashi, M, Ishikawa, K. | | Deposit date: | 2022-10-26 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of thermostable acetaldehyde dehydrogenase from the hyperthermophilic archaeon Sulfolobus tokodaii.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
3MKM
 
 | | Crystal structure of the E. coli pyrimidine nucleoside hydrolase YeiK (Apo-form) | | Descriptor: | CALCIUM ION, Putative uncharacterized protein YeiK | | Authors: | Garau, G, Fornili, A, Giabbai, B, Degano, M. | | Deposit date: | 2010-04-15 | | Release date: | 2010-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Energy Landscapes Associated with Macromolecular Conformational Changes from Endpoint Structures
J.Am.Chem.Soc., 132, 2010
|
|
2R2Z
 
 | | The crystal structure of a hemolysin domain from Enterococcus faecalis V583 | | Descriptor: | Hemolysin, ZINC ION | | Authors: | Zhang, R, Tan, K, Zhou, M, Bargassa, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-08-28 | | Release date: | 2007-09-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The crystal structure of a hemolysin domain from Enterococcus faecalis V583.
To be Published
|
|
