2FAR
 
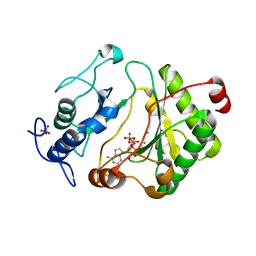 | | Crystal Structure of Pseudomonas aeruginosa LigD polymerase domain with dATP and Manganese | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Zhu, H, Nandakumar, J, Aniukwu, J, Wang, L.K, Glickman, M.S, Lima, C.D, Shuman, S. | | Deposit date: | 2005-12-07 | | Release date: | 2006-05-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Atomic structure and nonhomologous end-joining function of the polymerase component of bacterial DNA ligase D
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
5R11
 
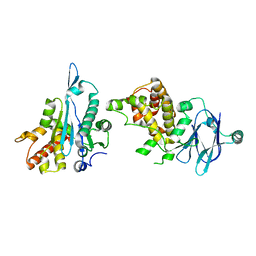 | | PanDDA analysis group deposition -- Auto-refined data of Aar2/RNaseH for ground state model 15, DMSO-free | | Descriptor: | A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R1J
 
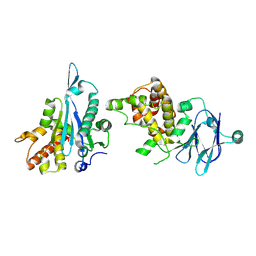 | | PanDDA analysis group deposition -- Auto-refined data of Aar2/RNaseH for ground state model 34, DMSO-free | | Descriptor: | A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-06-03 | | Last modified: | 2021-03-10 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R1X
 
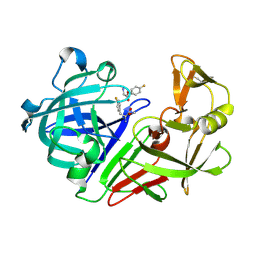 | | PanDDA analysis group deposition -- Endothiapepsin in complex with fragment F2X-Entry D04, DMSO-free | | Descriptor: | Endothiapepsin, methyl 4-fluoro-L-phenylalaninate | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (1.069 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R2C
 
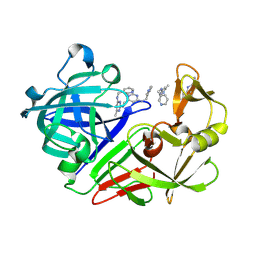 | | PanDDA analysis group deposition -- Endothiapepsin in complex with fragment F2X-Entry H05, DMSO-free | | Descriptor: | 6-methyl-N~4~-[(pyridin-3-yl)methyl]pyrimidine-2,4-diamine, Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (1.184 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R2S
 
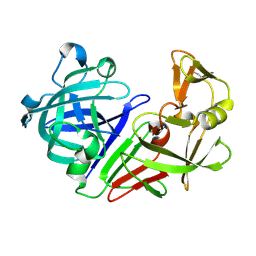 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 16, DMSO-Free | | Descriptor: | Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.149 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R37
 
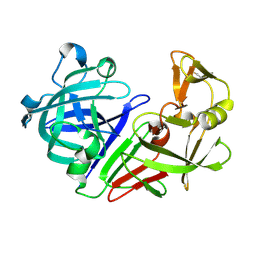 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 31, DMSO-Free | | Descriptor: | Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.038 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R3O
 
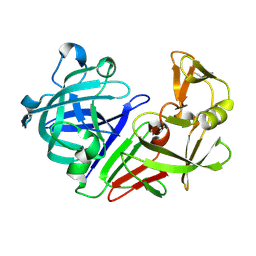 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 48, DMSO-Free | | Descriptor: | Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (1.217 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
7NEP
 
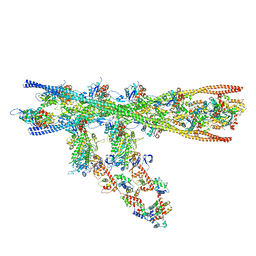 | | Homology model of the in situ actomyosin complex from the A-band of mouse psoas muscle sarcomere in the rigor state | | Descriptor: | Actin, alpha skeletal muscle, Myosin light chain 1/3, ... | | Authors: | Wang, Z, Grange, M, Wagner, T, Kho, A.L, Gautel, M, Raunser, S. | | Deposit date: | 2021-02-04 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (10.2 Å) | | Cite: | The molecular basis for sarcomere organization in vertebrate skeletal muscle.
Cell, 184, 2021
|
|
6YHP
 
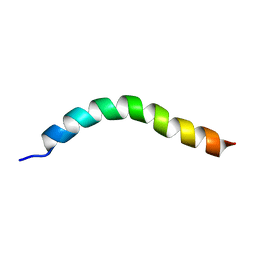 | | Solution NMR Structure of APP V44M mutant TMD | | Descriptor: | Amyloid-beta precursor protein V44M mutant | | Authors: | Silber, M, Muhle-Goll, C. | | Deposit date: | 2020-03-30 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Altered Hinge Conformations in APP Transmembrane Helix Mutants May Affect Enzyme-Substrate Interactions of gamma-Secretase.
Acs Chem Neurosci, 11, 2020
|
|
6YHX
 
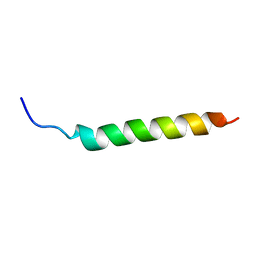 | | Solution NMR Structure of APP I45T mutant TMD | | Descriptor: | Amyloid-beta precursor protein I45T mutant | | Authors: | Silber, M, Muhle-Goll, C. | | Deposit date: | 2020-03-31 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Altered Hinge Conformations in APP Transmembrane Helix Mutants May Affect Enzyme-Substrate Interactions of gamma-Secretase.
Acs Chem Neurosci, 11, 2020
|
|
6VCC
 
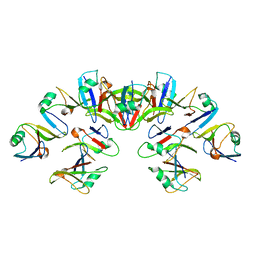 | | Cryo-EM structure of the Dvl2 DIX filament | | Descriptor: | Segment polarity protein dishevelled homolog DVL-2 | | Authors: | Enos, M, Kan, W, Muennich, S, Chen, D.H, Skiniotis, G, Weis, W.I. | | Deposit date: | 2019-12-20 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Limited Dishevelled/Axin oligomerization determines efficiency of Wnt/ beta-catenin signal transduction.
Elife, 9, 2020
|
|
8ORV
 
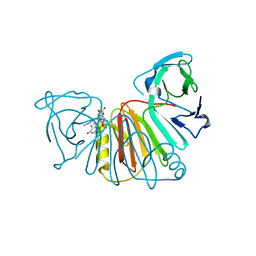 | | Crystal structure of monkeypox virus poxin in complex with the STING agonist MD1203 | | Descriptor: | 9-[(1~{S},6~{R},8~{R},9~{R},10~{R},15~{R},17~{R},18~{R})-8-(6-aminopurin-9-yl)-9,18-bis(fluoranyl)-3,12-bis(oxidanyl)-3,12-bis(oxidanylidene)-2,4,7,11,13-pentaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecan-17-yl]-1~{H}-purin-6-one, MPXVgp165 | | Authors: | Duchoslav, V, Klima, M, Boura, E. | | Deposit date: | 2023-04-17 | | Release date: | 2023-12-20 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Fluorinated cGAMP analogs, which act as STING agonists and are not cleavable by poxins: Structural basis of their function.
Structure, 32, 2024
|
|
6GBG
 
 | | Helicobacter pylori adhesin HopQ type I bound to the N-terminal domain of human CEACAM1 | | Descriptor: | BROMIDE ION, Carcinoembryonic antigen-related cell adhesion molecule 1, Outer membrane protein | | Authors: | Moonens, K, Kruse, T, Gerhard, M, Remaut, H. | | Deposit date: | 2018-04-13 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Helicobacter pyloriadhesin HopQ disruptstransdimerization in human CEACAMs.
EMBO J., 37, 2018
|
|
8P45
 
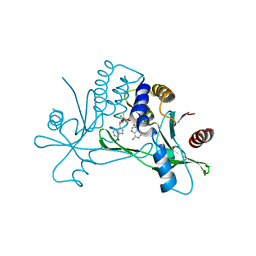 | | Crystal structure of human STING in complex with the agonist MD1202D | | Descriptor: | 9-[(1~{S},3~{R},6~{R},8~{R},9~{R},10~{R},12~{R},15~{R},17~{R},18~{R})-8-(6-aminopurin-9-yl)-9,18-bis(fluoranyl)-3,12-bis(oxidanylidene)-3,12-bis(sulfanyl)-2,4,7,11,13-pentaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecan-17-yl]-1~{H}-purin-6-one, Stimulator of interferon genes protein | | Authors: | Klima, M, Boura, E. | | Deposit date: | 2023-05-19 | | Release date: | 2023-12-20 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Fluorinated cGAMP analogs, which act as STING agonists and are not cleavable by poxins: Structural basis of their function.
Structure, 32, 2024
|
|
8ORW
 
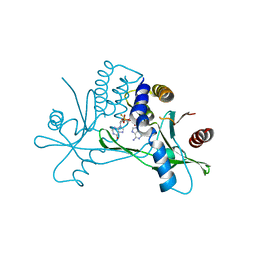 | | Crystal structure of human STING in complex with the agonist MD1203 | | Descriptor: | 9-[(1~{S},6~{R},8~{R},9~{R},10~{R},15~{R},17~{R},18~{R})-8-(6-aminopurin-9-yl)-9,18-bis(fluoranyl)-3,12-bis(oxidanyl)-3,12-bis(oxidanylidene)-2,4,7,11,13-pentaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecan-17-yl]-1~{H}-purin-6-one, Stimulator of interferon protein | | Authors: | Klima, M, Boura, E. | | Deposit date: | 2023-04-17 | | Release date: | 2023-12-20 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Fluorinated cGAMP analogs, which act as STING agonists and are not cleavable by poxins: Structural basis of their function.
Structure, 32, 2024
|
|
5R17
 
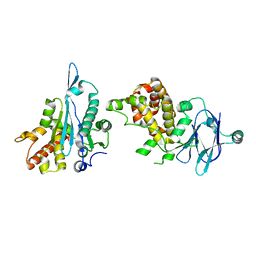 | | PanDDA analysis group deposition -- Auto-refined data of Aar2/RNaseH for ground state model 22, DMSO-free | | Descriptor: | A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
8P44
 
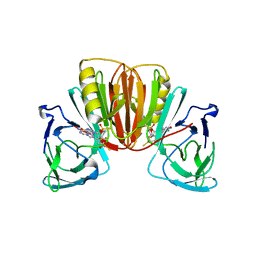 | | Crystal structure of monkeypox virus poxin in complex with the STING agonist MD1202D | | Descriptor: | 9-[(1~{S},3~{R},6~{R},8~{R},9~{R},10~{R},12~{R},15~{R},17~{R},18~{R})-8-(6-aminopurin-9-yl)-9,18-bis(fluoranyl)-3,12-bis(oxidanylidene)-3,12-bis(sulfanyl)-2,4,7,11,13-pentaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecan-17-yl]-1~{H}-purin-6-one, MPXVgp165 | | Authors: | Duchoslav, V, Klima, M, Boura, E. | | Deposit date: | 2023-05-19 | | Release date: | 2023-12-20 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Fluorinated cGAMP analogs, which act as STING agonists and are not cleavable by poxins: Structural basis of their function.
Structure, 32, 2024
|
|
5R1M
 
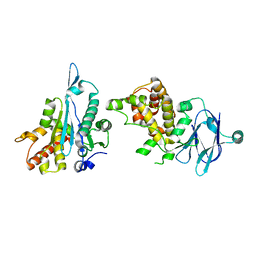 | | PanDDA analysis group deposition -- Auto-refined data of Aar2/RNaseH for ground state model 37, DMSO-free | | Descriptor: | A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R23
 
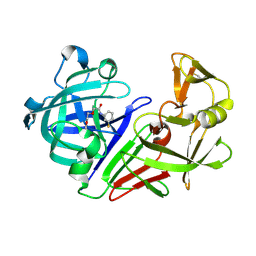 | | PanDDA analysis group deposition -- Endothiapepsin in complex with fragment F2X-Entry F03, DMSO-free | | Descriptor: | Endothiapepsin, N-(3-fluorophenyl)-2-(2-methoxyethoxy)acetamide | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.029 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R2F
 
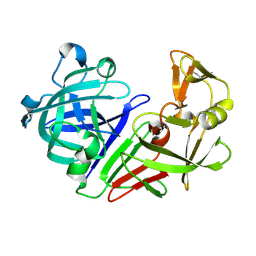 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 02, DMSO-Free | | Descriptor: | Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.119 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R2V
 
 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 19, DMSO-Free | | Descriptor: | Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.048 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R3C
 
 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 36, DMSO-Free | | Descriptor: | Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (0.949 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R3Q
 
 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 50, DMSO-Free | | Descriptor: | Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (1.009 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5RC1
 
 | | PanDDA analysis group deposition -- Endothiapepsin changed state model for fragment F2X-Entry Library E07a | | Descriptor: | 1-(1-methyl-1,2,3,4-tetrahydroquinolin-6-yl)methanamine, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
