6MJW
 
 | | human cGAS catalytic domain bound with the inhibitor G150 | | Descriptor: | 1-[9-(6-aminopyridin-3-yl)-6,7-dichloro-1,3,4,5-tetrahydro-2H-pyrido[4,3-b]indol-2-yl]-2-hydroxyethan-1-one, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Lama, L, Adura, C, Xie, W, Tomita, D, Kamei, T, Kuryavyi, V, Gogakos, T, Steinberg, J.I, Miller, M, Ramos-Espiritu, L, Asano, Y, Hashizume, S, Aida, J, Imaeda, T, Okamoto, R, Jennings, A.J, Michinom, M, Kuroita, T, Stamford, A, Gao, P, Meinke, P, Glickman, J.F, Patel, D.J, Tuschl, T. | | Deposit date: | 2018-09-23 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.405 Å) | | Cite: | Development of human cGAS-specific small-molecule inhibitors for repression of dsDNA-triggered interferon expression.
Nat Commun, 10, 2019
|
|
8DG9
 
 | |
8DG8
 
 | |
3GGP
 
 | | Crystal structure of prephenate dehydrogenase from A. aeolicus in complex with hydroxyphenyl propionate and NAD+ | | Descriptor: | CHLORIDE ION, HYDROXYPHENYL PROPIONIC ACID, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Sun, W, Shahinas, D, Kimber, M.S, Christendat, D. | | Deposit date: | 2009-03-01 | | Release date: | 2009-03-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Crystal Structure of Aquifex aeolicus Prephenate Dehydrogenase Reveals the Mode of Tyrosine Inhibition.
J.Biol.Chem., 284, 2009
|
|
6Y07
 
 | |
6Y2K
 
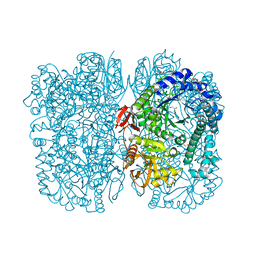 | | Crystal structure of beta-galactosidase from the psychrophilic Marinomonas ef1 | | Descriptor: | CHLORIDE ION, GLYCEROL, beta-galactosidase | | Authors: | Mangiagalli, M, Lapi, M, Maione, S, Orlando, M, Brocca, S, Pesce, A, Barbiroli, A, Pucciarelli, S, Camilloni, C, Lotti, M. | | Deposit date: | 2020-02-16 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The co-existence of cold activity and thermal stability in an Antarctic GH42 beta-galactosidase relies on its hexameric quaternary arrangement.
Febs J., 288, 2021
|
|
6Y4C
 
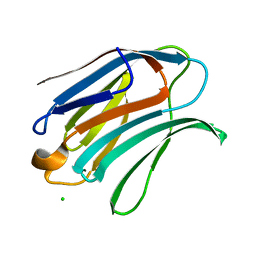 | | Structure of galectin-3C in complex with lactose determined by serial crystallography using an XtalTool support | | Descriptor: | CHLORIDE ION, Galectin-3, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Shilova, A, Hakansson, M, Welin, M, Kovacic, R, Mueller, U, Logan, D.T. | | Deposit date: | 2020-02-20 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Current status and future opportunities for serial crystallography at MAX IV Laboratory.
J.Synchrotron Radiat., 27, 2020
|
|
3FSY
 
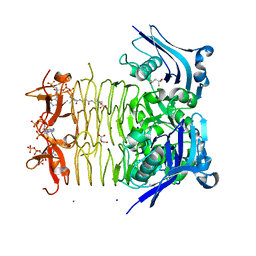 | | Structure of tetrahydrodipicolinate N-succinyltransferase (Rv1201c;DapD) in complex with succinyl-CoA from Mycobacterium tuberculosis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, MAGNESIUM ION, ... | | Authors: | Schuldt, L, Weyand, S, Kefala, G, Weiss, M.S. | | Deposit date: | 2009-01-12 | | Release date: | 2009-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The three-dimensional Structure of a mycobacterial DapD provides insights into DapD diversity and reveals unexpected particulars about the enzymatic mechanism.
J.Mol.Biol., 389, 2009
|
|
6GA8
 
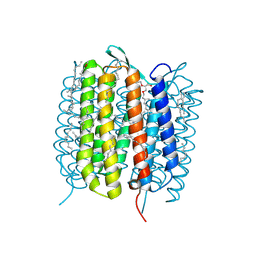 | | BACTERIORHODOPSIN, 330 FS STATE, REAL-SPACE REFINED AGAINST 15% EXTRAPOLATED STRUCTURE FACTORS | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nass Kovacs, G, Colletier, J.-P, Gruenbein, M.L, Stensitzki, T, Batyuk, A, Carbajo, S, Doak, R.B, Ehrenberg, D, Foucar, L, Gasper, R, Gorel, A, Hilpert, M, Kloos, M, Koglin, J, Reinstein, J, Roome, C.M, Schlesinger, R, Seaberg, M, Shoeman, R.L, Stricker, M, Boutet, S, Haacke, S, Heberle, J, Domratcheva, T, Schlichting, I. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin.
Nat Commun, 10, 2019
|
|
6GAH
 
 | | BACTERIORHODOPSIN, 680 FS STATE, REAL-SPACE REFINED AGAINST 15% EXTRAPOLATED STRUCTURE FACTORS | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nass Kovacs, G, Colletier, J.-P, Gruenbein, M.L, Stensitzki, T, Batyuk, A, Carbajo, S, Doak, R.B, Ehrenberg, D, Foucar, L, Gasper, R, Gorel, A, Hilpert, M, Kloos, M, Koglin, J, Reinstein, J, Roome, C.M, Schlesinger, R, Seaberg, M, Shoeman, R.L, Stricker, M, Boutet, S, Haacke, S, Heberle, J, Domratcheva, T, Schlichting, I. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin.
Nat Commun, 10, 2019
|
|
7NCJ
 
 | |
4ZEA
 
 | | Endothiapepsin in complex with fragment B91 | | Descriptor: | 1,2-ETHANEDIOL, 2-IMINOBIOTIN, ACETATE ION, ... | | Authors: | Huschmann, F.U, Linnik, J, Weiss, M.S, Mueller, U. | | Deposit date: | 2015-04-20 | | Release date: | 2016-05-04 | | Last modified: | 2016-05-11 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structures of endothiapepsin-fragment complexes from crystallographic fragment screening using a novel, diverse and affordable 96-compound fragment library.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
7NCG
 
 | |
2F47
 
 | | Xray crystal structure of T4 lysozyme mutant L20/R63A liganded to methylguanidinium | | Descriptor: | 1-METHYLGUANIDINE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Yousef, M.S, Bischoff, N, Dyer, C.M, Baase, W.A, Matthews, B.W. | | Deposit date: | 2005-11-22 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Guanidinium derivatives bind preferentially and trigger long-distance conformational changes in an engineered T4 lysozyme.
Protein Sci., 15, 2006
|
|
7NCI
 
 | |
7NCH
 
 | |
7NCK
 
 | |
7NCA
 
 | |
6GA7
 
 | | BACTERIORHODOPSIN, 240FS STATE, REAL-SPACE REFINED AGAINST 10% EXTRAPOLATED MAP | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nass Kovacs, G, Colletier, J.-P, Gruenbein, M.L, Stensitzki, T, Batyuk, A, Carbajo, S, Doak, R.B, Ehrenberg, D, Foucar, L, Gasper, R, Gorel, A, Hilpert, M, Kloos, M, Koglin, J, Reinstein, J, Roome, C.M, Schlesinger, R, Seaberg, M, Shoeman, R.L, Stricker, M, Boutet, S, Haacke, S, Heberle, J, Domratcheva, T, Schlichting, I. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin.
Nat Commun, 10, 2019
|
|
6GAI
 
 | | BACTERIORHODOPSIN, 740 FS STATE, REAL-SPACE REFINED AGAINST 15% EXTRAPOLATED STRUCTURE FACTORS | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nass Kovacs, G, Colletier, J.-P, Gruenbein, M.L, Stensitzki, T, Batyuk, A, Carbajo, S, Doak, R.B, Ehrenberg, D, Foucar, L, Gasper, R, Gorel, A, Hilpert, M, Kloos, M, Koglin, J, Reinstein, J, Roome, C.M, Schlesinger, R, Seaberg, M, Shoeman, R.L, Stricker, M, Boutet, S, Haacke, S, Heberle, J, Domratcheva, T, Schlichting, I. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin.
Nat Commun, 10, 2019
|
|
6P8M
 
 | |
4ZOP
 
 | | Co-crystal Structure of Lipid Kinase PI3K alpha with a selective phosphatidylinositol-3 kinase alpha inhibitor | | Descriptor: | (2S,3R)-N~1~-(8-tert-butyl-4,5-dihydro[1,3]thiazolo[4,5-h]quinazolin-2-yl)-3-methylpyrrolidine-1,2-dicarboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Knapp, M.S, Elling, R.A. | | Deposit date: | 2015-05-06 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Co-crystal Structure of the Lipid Kinase PI3K alpha with a selective phosphatidylinositol-3 kinase alpha inhibitorCo-crystal Structure of the Lipid Kinase PI3K alpha with a selective phosphatidylinositol-3 kinase alpha inhibitor
To Be Published
|
|
6CAE
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with NOSO-95179 antibiotic and bound to mRNA and A-, P- and E-site tRNAs at 2.6A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Pantel, L, Florin, T, Dobosz-Bartoszek, M, Racine, E, Sarciaux, M, Serri, M, Houard, J, Campagne, J.M, Marcia de Figueiredo, R, Midrier, C, Gaudriault, S, Givaudan, A, Lanois, A, Forst, S, Aumelas, A, Cotteaux-Lautard, C, Bolla, J.M, Vingsbo Lundberg, C, Huseby, D, Hughes, D, Villain-Guillot, P, Mankin, A.S, Polikanov, Y.S, Gualtieri, M. | | Deposit date: | 2018-01-30 | | Release date: | 2018-04-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Odilorhabdins, Antibacterial Agents that Cause Miscoding by Binding at a New Ribosomal Site.
Mol. Cell, 70, 2018
|
|
3FSX
 
 | | Structure of tetrahydrodipicolinate N-succinyltransferase (Rv1201c; DapD) from Mycobacterium tuberculosis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, MAGNESIUM ION, ... | | Authors: | Schuldt, L, Weyand, S, Kefala, G, Weiss, M.S. | | Deposit date: | 2009-01-12 | | Release date: | 2009-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The three-dimensional Structure of a mycobacterial DapD provides insights into DapD diversity and reveals unexpected particulars about the enzymatic mechanism.
J.Mol.Biol., 389, 2009
|
|
6GA6
 
 | | Bacteriorhodopsin, 10 ps state, real-space refined against 10% extrapolated map | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nass Kovacs, G, Colletier, J.-P, Gruenbein, M.L, Stensitzki, T, Batyuk, A, Carbajo, S, Doak, R.B, Ehrenberg, D, Foucar, L, Gasper, R, Gorel, A, Hilpert, M, Kloos, M, Koglin, J, Reinstein, J, Roome, C.M, Schlesinger, R, Seaberg, M, Shoeman, R.L, Stricker, M, Boutet, S, Haacke, S, Heberle, J, Domratcheva, T, Schlichting, I. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin.
Nat Commun, 10, 2019
|
|
