8F0R
 
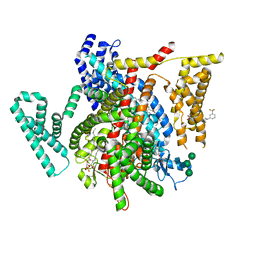 | | Structure of VSD4-NaV1.7-NaVPas channel chimera bound to the arylsulfonamide inhibitor GNE-3565 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-chloro-4-({(1S,2S,4S)-2-(dimethylamino)-4-[3-(trifluoromethyl)phenyl]cyclohexyl}amino)-2-fluoro-N-(pyrimidin-4-yl)benzene-1-sulfonamide, ... | | Authors: | Kschonsak, M, Jao, C.C, Arthur, C.P, Rohou, A.L, Bergeron, P, Ortwine, D, McKerall, S.J, Hackos, D.H, Deng, L, Chen, J, Sutherlin, D, Dragovich, P.S, Volgraf, M, Wright, M.R, Payandeh, J, Ciferri, C, Tellis, J.C. | | Deposit date: | 2022-11-03 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM reveals an unprecedented binding site for Na V 1.7 inhibitors enabling rational design of potent hybrid inhibitors.
Elife, 12, 2023
|
|
8CC2
 
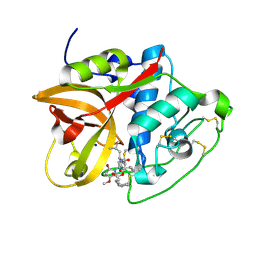 | | Cathepsin B1 from Schistosoma mansoni in complex with gallinamide A | | Descriptor: | Cathepsin B-like peptidase (C01 family), SODIUM ION, gallinamide A, ... | | Authors: | Rubesova, P, Brynda, J, Mares, M, Gerwick, W.H. | | Deposit date: | 2023-01-26 | | Release date: | 2024-02-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Nature-Inspired Gallinamides Are Potent Antischistosomal Agents: Inhibition of the Cathepsin B1 Protease Target and Binding Mode Analysis.
Acs Infect Dis., 10, 2024
|
|
6BMW
 
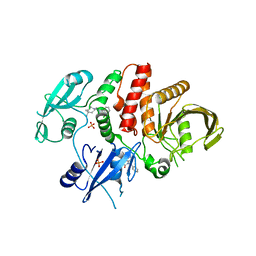 | | Non-receptor Protein Tyrosine Phosphatase SHP2 in Complex with Allosteric Inhibitors SHP099 and SHP504 | | Descriptor: | 3-{4-[(2-chlorophenyl)methyl]-5-oxo-4,5-dihydro[1,2,4]triazolo[4,3-a]quinazolin-1-yl}-4-hydroxybenzoic acid, 6-(4-azanyl-4-methyl-piperidin-1-yl)-3-[2,3-bis(chloranyl)phenyl]pyrazin-2-amine, GLYCEROL, ... | | Authors: | Stams, T, Fodor, M. | | Deposit date: | 2017-11-15 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dual Allosteric Inhibition of SHP2 Phosphatase.
ACS Chem. Biol., 13, 2018
|
|
8CCU
 
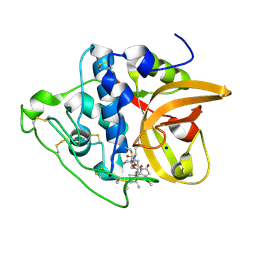 | | Cathepsin B1 from Schistosoma mansoni in complex with gallinamide analog 1 | | Descriptor: | Cathepsin B-like peptidase (C01 family), SODIUM ION, [(2~{S})-1-[[(2~{S})-1-[[(2~{S})-5-[(2~{S})-3-methoxy-2-(2-methylpropyl)-5-oxidanylidene-2~{H}-pyrrol-1-yl]-5-oxidanylidene-pentan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl] (2~{S},3~{S})-2-(dimethylamino)-3-methyl-pentanoate | | Authors: | Rubesova, P, Brynda, J, Gerwick, W.H, Mares, M. | | Deposit date: | 2023-01-27 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Nature-Inspired Gallinamides Are Potent Antischistosomal Agents: Inhibition of the Cathepsin B1 Protease Target and Binding Mode Analysis.
Acs Infect Dis., 10, 2024
|
|
4XRH
 
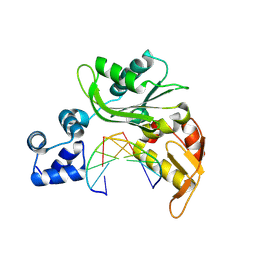 | |
6BRB
 
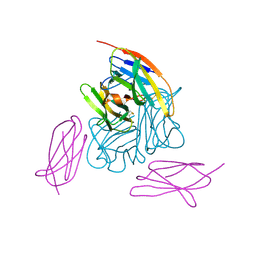 | | Novel non-antibody protein scaffold targeting CD40L | | Descriptor: | CD40 ligand, Tn3-like, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Oganesyan, V, Baca, M, Thisted, T, Grinberg, L, Wu, H, Dall'Acqua, W.F. | | Deposit date: | 2017-11-30 | | Release date: | 2018-12-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | A CD40L-targeting protein reduces autoantibodies and improves disease activity in patients with autoimmunity.
Sci Transl Med, 11, 2019
|
|
8C8A
 
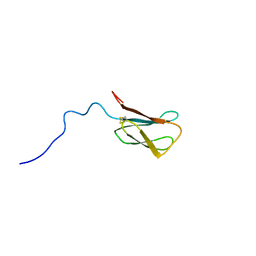 | | The NMR structure of the MAX28 effector from Magnaporthe oryzae | | Descriptor: | But2 domain-containing protein | | Authors: | Lahfa, M, Padilla, A, de Guillen, K, Kroj, T, Roumestand, C, Barthe, P. | | Deposit date: | 2023-01-19 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-04 | | Method: | SOLUTION NMR | | Cite: | The structural landscape and diversity of Pyricularia oryzae MAX effectors revisited.
Plos Pathog., 20, 2024
|
|
7Q3B
 
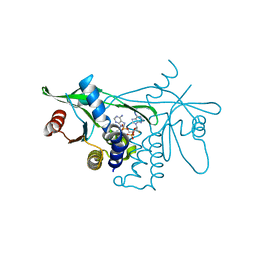 | | Crystal structure of human STING in complex with 3'3'-c-(2'F,2'dA-isonucA)MP | | Descriptor: | 3'3'-c-(2'F,2'dA-isonucA)MP, Stimulator of interferon genes protein | | Authors: | Smola, M, Klima, M, Boura, E. | | Deposit date: | 2021-10-27 | | Release date: | 2022-06-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.55733919 Å) | | Cite: | Discovery of isonucleotidic CDNs as potent STING agonists with immunomodulatory potential.
Structure, 30, 2022
|
|
6D2L
 
 | | Crystal structure of human CARM1 with (S)-SKI-72 | | Descriptor: | (2S,5S)-2-amino-6-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]-5-[(benzylamino)methyl]-N-[2-(4-hydroxyphenyl)ethyl]hexanamide, GLYCEROL, Histone-arginine methyltransferase CARM1, ... | | Authors: | DONG, A, ZENG, H, WALKER, J.R, Hutchinson, A, Seitova, A, LUO, M, CAI, X.C, KE, W, WANG, J, SHI, C, ZHENG, W, LEE, J.P, IBANEZ, G, Bountra, C, Arrowsmith, C.H, Edwards, A.M, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-04-13 | | Release date: | 2018-05-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A chemical probe of CARM1 alters epigenetic plasticity against breast cancer cell invasion.
Elife, 8, 2019
|
|
6CBE
 
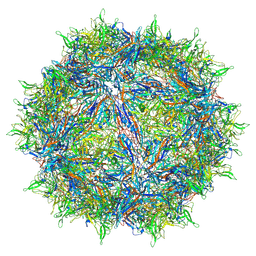 | | Atomic structure of a rationally engineered gene delivery vector, AAV2.5 | | Descriptor: | Capsid protein VP1 | | Authors: | Burg, M, Rosebrough, C, Drouin, L, Bennett, A, Mietzsch, M, Chipman, P, McKenna, R, Sousa, D, Potter, M, Byrne, B, Kozyreva, O.G, Samulski, R.J, Agbandje-McKenna, M. | | Deposit date: | 2018-02-02 | | Release date: | 2018-05-30 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Atomic structure of a rationally engineered gene delivery vector, AAV2.5.
J. Struct. Biol., 203, 2018
|
|
4XQ8
 
 | |
8BZU
 
 | |
6ZRW
 
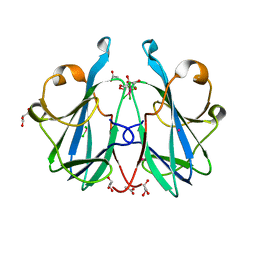 | | Crystal structure of the fungal lectin CML1 | | Descriptor: | ACETATE ION, GLYCEROL, Mucin-binding lectin 1, ... | | Authors: | Bleuler-Martinez, S, Olieric, V, Sharpe, M, Capitani, G, Aebi, M, Kuenzler, M. | | Deposit date: | 2020-07-15 | | Release date: | 2021-07-28 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure-function relationship of a novel fucoside-binding fruiting body lectin from Coprinopsis cinerea exhibiting nematotoxic activity.
Glycobiology, 32, 2022
|
|
4Y0B
 
 | | The structure of Arabidopsis ClpT1 | | Descriptor: | CHLORIDE ION, Double Clp-N motif protein | | Authors: | Kimber, M.S, Schultz, L. | | Deposit date: | 2015-02-05 | | Release date: | 2015-05-13 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures, Functions, and Interactions of ClpT1 and ClpT2 in the Clp Protease System of Arabidopsis Chloroplasts.
Plant Cell, 27, 2015
|
|
4Y38
 
 | | Endothiapepsin in complex with fragment B29 | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huschmann, F.U, Linnik, J, Weiss, M.S, Mueller, U. | | Deposit date: | 2015-02-10 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structures of endothiapepsin-fragment complexes from crystallographic fragment screening using a novel, diverse and affordable 96-compound fragment library.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
6P8N
 
 | |
6FGA
 
 | | Crystal structure of TRIM21 E3 ligase, RING domain in complex with its cognate E2 conjugating enzyme UBE2E1 | | Descriptor: | E3 ubiquitin-protein ligase TRIM21, GLYCEROL, Ubiquitin-conjugating enzyme E2 E1, ... | | Authors: | Anandapadamanaban, M, Moche, M, Sunnerhagen, M. | | Deposit date: | 2018-01-10 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | E3 ubiquitin-protein ligase TRIM21-mediated lysine capture by UBE2E1 reveals substrate-targeting mode of a ubiquitin-conjugating E2.
J.Biol.Chem., 294, 2019
|
|
6XYP
 
 | | Multiple system atrophy Type II-1 alpha-synuclein filament | | Descriptor: | Alpha-synuclein | | Authors: | Schweighauser, M, Shi, Y, Tarutani, A, Kametani, F, Murzin, A.G, Ghetti, B, Matsubara, T, Tomita, T, Ando, T, Hasegawa, K, Murayama, S, Yoshida, M, Hasegawa, M, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2020-01-30 | | Release date: | 2020-02-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structures of alpha-synuclein filaments from multiple system atrophy.
Nature, 585, 2020
|
|
6CHM
 
 | |
4F4C
 
 | | The Crystal Structure of the Multi-Drug Transporter | | Descriptor: | Multidrug resistance protein pgp-1, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, undecyl 4-O-alpha-D-glucopyranosyl-1-thio-beta-D-glucopyranoside | | Authors: | Jin, M.S, Oldham, M.L, Zhang, Q, Chen, J. | | Deposit date: | 2012-05-10 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of the multidrug transporter P-glycoprotein from Caenorhabditis elegans.
Nature, 490, 2012
|
|
6U9U
 
 | | Structure of GM9_TH8seq732127 FAB | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, GM9_TH8seq732127 FAB heavy chain, ... | | Authors: | Singh, S, Liban, T.J, Pancera, M. | | Deposit date: | 2019-09-09 | | Release date: | 2019-11-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Extensive dissemination and intraclonal maturation of HIV Env vaccine-induced B cell responses.
J.Exp.Med., 217, 2020
|
|
6XYQ
 
 | | Multiple system atrophy Type II-2 alpha-synuclein filament | | Descriptor: | Alpha-synuclein | | Authors: | Schweighauser, M, Shi, Y, Tarutani, A, Kametani, F, Murzin, A.G, Ghetti, B, Matsubara, T, Tomita, T, Ando, T, Hasegawa, K, Murayama, S, Yoshida, M, Hasegawa, M, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2020-01-30 | | Release date: | 2020-02-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structures of alpha-synuclein filaments from multiple system atrophy.
Nature, 585, 2020
|
|
6PEH
 
 | | Crystal structure of rabbit monoclonal anti-HIV antibody 1C2 | | Descriptor: | 1C2 Fab Heavy Chain, 1C2 Fab Light Chain, SULFATE ION | | Authors: | Liban, T, Pancera, M. | | Deposit date: | 2019-06-20 | | Release date: | 2019-11-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.296 Å) | | Cite: | Vaccination with Glycan-Modified HIV NFL Envelope Trimer-Liposomes Elicits Broadly Neutralizing Antibodies to Multiple Sites of Vulnerability.
Immunity, 51, 2019
|
|
4XPL
 
 | | The crystal structure of Campylobacter jejuni N-acetyltransferase PseH in complex with acetyl coenzyme A | | Descriptor: | ACETYL COENZYME *A, N-Acetyltransferase, PseH | | Authors: | Song, W.S, Nam, M.S, Namgung, B, Yoon, S.I. | | Deposit date: | 2015-01-17 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analysis of PseH, the Campylobacter jejuni N-acetyltransferase involved in bacterial O-linked glycosylation.
Biochem.Biophys.Res.Commun., 458, 2015
|
|
5JOH
 
 | | CRYSTAL STRUCTURE OF CSN5(2-257) IN COMPLEX WITH CNS5i-1b | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2'-chloro-6-[(5S,6S)-6-hydroxy-6,7,8,9-tetrahydro-5H-imidazo[1,5-a]azepin-5-yl][1,1'-biphenyl]-3-carbonitrile, COP9 signalosome complex subunit 5, ... | | Authors: | Renatus, M, Wiesmann, C. | | Deposit date: | 2016-05-02 | | Release date: | 2016-11-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Targeted inhibition of the COP9 signalosome for treatment of cancer.
Nat Commun, 7, 2016
|
|
