3B59
 
 | |
7EGU
 
 | | Structure of human NNMT in complex with macrocyclic peptide X | | Descriptor: | Nicotinamide N-methyltransferase, macrocyclic peptide X | | Authors: | Hayashi, K, Mikamiyama, H, Uehara, S, Yamamoto, S, Cary, D, Nishikawa, J, Ueda, T, Ozasa, H, Mihara, K, Yoshimura, N, Kawai, T, Ono, T, Yamamoto, S, Fumoto, M. | | Deposit date: | 2021-03-26 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Macrocyclic Peptides as a Novel Class of NNMT Inhibitors: A SAR Study Aimed at Inhibitory Activity in the Cell.
Acs Med.Chem.Lett., 12, 2021
|
|
3B7H
 
 | |
3H5H
 
 | |
3BA6
 
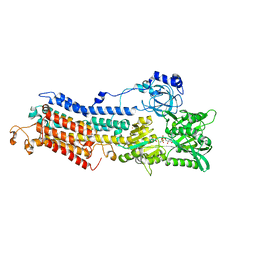 | | Structure of the Ca2E1P phosphoenzyme intermediate of the SERCA Ca2+-ATPase | | Descriptor: | AMP PHOSPHORAMIDATE, CALCIUM ION, POTASSIUM ION, ... | | Authors: | Picard, M, Winther, A.M.L, Olesen, C, Gyrup, C, Morth, J.P, Oxvig, C, Moller, J.V, Nissen, P. | | Deposit date: | 2007-11-07 | | Release date: | 2007-12-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structural basis of calcium transport by the calcium pump.
Nature, 450, 2007
|
|
3BGE
 
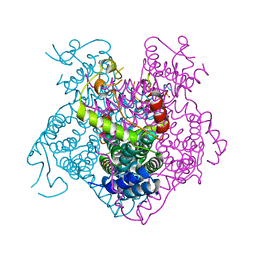 | | Crystal structure of the C-terminal fragment of AAA+ATPase from Haemophilus influenzae | | Descriptor: | Predicted ATPase, SULFATE ION | | Authors: | Ramagopal, U.A, Patskovsky, Y, Bonanno, J.B, Meyer, A.J, Toro, R, Freeman, J, Adams, J, Koss, J, Maletic, M, Gheyi, T, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-11-26 | | Release date: | 2008-01-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the C-terminal fragment of AAA+ATPase from Haemophilus influenzae.
To be Published
|
|
3BIK
 
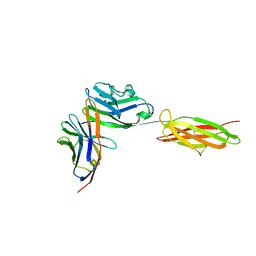 | | Crystal Structure of the PD-1/PD-L1 Complex | | Descriptor: | GLYCEROL, Programmed cell death 1 ligand 1, Programmed cell death protein 1 | | Authors: | Lin, D.Y, Tanaka, Y, Iwasaki, M, Gittis, A.G, Su, H.P, Mikami, B, Okazaki, T, Honjo, T, Minato, N, Garboczi, D.N. | | Deposit date: | 2007-11-30 | | Release date: | 2008-02-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The PD-1/PD-L1 complex resembles the antigen-binding Fv domains of antibodies and T cell receptors.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3BFH
 
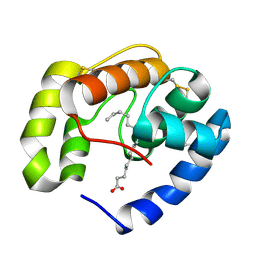 | | Crystal structure of a pheromone binding protein from Apis mellifera in complex with hexadecanoic acid | | Descriptor: | CHLORIDE ION, PALMITIC ACID, Pheromone-binding protein ASP1 | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2007-11-21 | | Release date: | 2008-06-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the honey bee PBP pheromone and pH-induced conformational change
J.Mol.Biol., 380, 2008
|
|
7WFD
 
 | | Left PSI in the cyclic electron transport supercomplex NDH-PSI from Arabidopsis | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Pan, X, Li, M. | | Deposit date: | 2021-12-26 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Supramolecular assembly of chloroplast NADH dehydrogenase-like complex with photosystem I from Arabidopsis thaliana.
Mol Plant, 15, 2022
|
|
6KF9
 
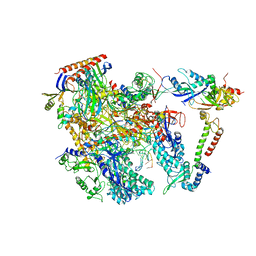 | | Cryo-EM structure of Thermococcus kodakarensis RNA polymerase | | Descriptor: | DNA (27-MER), DNA (5'-D(P*TP*CP*GP*GP*TP*AP*AP*TP*CP*AP*CP*GP*CP*TP*CP*C)-3'), DNA-directed RNA polymerase subunit, ... | | Authors: | Jun, S.-H, Hyun, J, Jeong, H, Cha, J.S, Kim, H, Bartlett, M.S, Cho, H.-S, Murakami, K.S. | | Deposit date: | 2019-07-07 | | Release date: | 2020-07-01 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Direct binding of TFE alpha opens DNA binding cleft of RNA polymerase.
Nat Commun, 11, 2020
|
|
3B4O
 
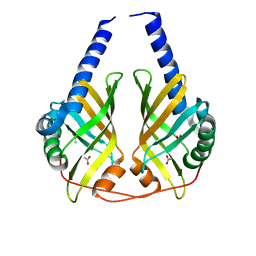 | | Crystal structure of phenazine biosynthesis protein PhzA/B from Burkholderia cepacia R18194, apo form | | Descriptor: | ACETATE ION, Phenazine biosynthesis protein A/B | | Authors: | Ahuja, E.G, Janning, P, Mentel, M, Graebsch, A, Breinbauer, R, Blankenfeldt, W. | | Deposit date: | 2007-10-24 | | Release date: | 2008-12-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | PhzA/B catalyzes the formation of the tricycle in phenazine biosynthesis.
J.Am.Chem.Soc., 130, 2008
|
|
3KIC
 
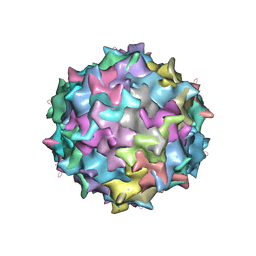 | |
3B57
 
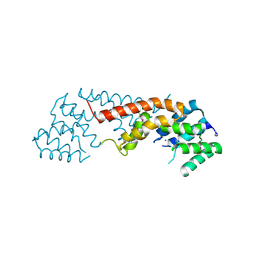 | | Crystal structure of the Lin1889 protein (Q92AN1) from Listeria innocua. Northeast Structural Consortium target LkR65 | | Descriptor: | Lin1889 protein, MAGNESIUM ION | | Authors: | Kuzin, A.P, Abashidze, M, Seetharaman, J, Benach, J, Forouhar, F, Vorobiev, S.M, Wang, H, Janjua, H, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-10-25 | | Release date: | 2007-11-06 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the Lin1889 protein (Q92AN1) from Listeria innocua.
To be Published
|
|
3B2C
 
 | | Crystal structure of the collagen triple helix model [{PRO-HYP(R)-GLY}4-{HYP(S)-Pro-GLY}2-{PRO-HYP(R)-GLY}4]3 | | Descriptor: | Collagen-like peptide | | Authors: | Motooka, D, Kawahara, K, Nakamura, S, Doi, M, Nishi, Y, Nishiuchi, Y, Nakazawa, T, Yoshida, T, Ohkubo, T, Kobayashi, Y, Kang, Y.K, Uchiyama, S. | | Deposit date: | 2011-07-26 | | Release date: | 2012-04-04 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | The triple helical structure and stability of collagen model peptide with 4(S)-hydroxyprolyl-pro-gly units
Biopolymers, 98, 2011
|
|
3B85
 
 | |
7W2V
 
 | | Crystal structure of TxGH116 R786A mutant from Thermoanaerobacterium xylanolyticum with glucose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Huang, M, Pengthaisong, S, Charoenwattanasatien, R, Jitonnom, J, Ketudat Cairns, J.R. | | Deposit date: | 2021-11-24 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Systematic Functional and Computational Analysis of Glucose-Binding Residues in Glycoside Hydrolase Family GH116.
Catalysts, 12, 2022
|
|
7W2S
 
 | | Crystal structure of TxGH116 E730A mutant from Thermoanaerobacterium xylanolyticum with glucose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Huang, M, Pengthaisong, S, Charoenwattanasatien, R, Jitonnom, J, Ketudat Cairns, J.R. | | Deposit date: | 2021-11-24 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Systematic Functional and Computational Analysis of Glucose-Binding Residues in Glycoside Hydrolase Family GH116.
Catalysts, 12, 2022
|
|
7DXO
 
 | |
7W2W
 
 | | Crystal structure of TxGH116 R786K mutant from Thermoanaerobacterium xylanolyticum with glucose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Huang, M, Pengthaisong, S, Charoenwattanasatien, R, Jitonnom, J, Ketudat Cairns, J.R. | | Deposit date: | 2021-11-24 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Systematic Functional and Computational Analysis of Glucose-Binding Residues in Glycoside Hydrolase Family GH116.
Catalysts, 12, 2022
|
|
7W2T
 
 | | Crystal structure of TxGH116 E730Q mutant from Thermoanaerobacterium xylanolyticum with glucose | | Descriptor: | CALCIUM ION, GLYCEROL, Glucosylceramidase, ... | | Authors: | Huang, M, Pengthaisong, S, Charoenwattanasatien, R, Jitonnom, J, Ketudat Cairns, J.R. | | Deposit date: | 2021-11-24 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Systematic Functional and Computational Analysis of Glucose-Binding Residues in Glycoside Hydrolase Family GH116.
Catalysts, 12, 2022
|
|
7DY3
 
 | |
3B6Y
 
 | | Crystal Structure of the Second HIN-200 Domain of Interferon-Inducible Protein 16 | | Descriptor: | Gamma-interferon-inducible protein Ifi-16, SULFATE ION | | Authors: | Liao, J.C.C, Lam, R, Ravichandran, M, Duan, S, Tempel, W, Chirgadze, N.Y, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-10-29 | | Release date: | 2007-11-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure Analysis of the Second HIN Domain of IFI16.
To be Published
|
|
7W2X
 
 | | Crystal structure of TxGH116 R786K mutant from Thermoanaerobacterium xylanolyticum | | Descriptor: | CALCIUM ION, GLYCEROL, Glucosylceramidase, ... | | Authors: | Huang, M, Pengthaisong, S, Charoenwattanasatien, R, Jitonnom, J, Ketudat Cairns, J.R. | | Deposit date: | 2021-11-24 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Systematic Functional and Computational Analysis of Glucose-Binding Residues in Glycoside Hydrolase Family GH116.
Catalysts, 12, 2022
|
|
3B7R
 
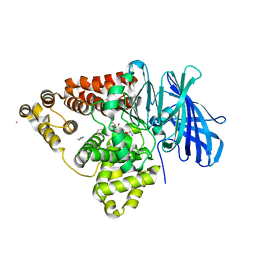 | | Leukotriene A4 Hydrolase Complexed with Inhibitor RB3040 | | Descriptor: | IMIDAZOLE, Leukotriene A-4 hydrolase, N-[3-[(1-AMINOETHYL)(HYDROXY)PHOSPHORYL]-2-(1,1'-BIPHENYL-4-YLMETHYL)PROPANOYL]ALANINE, ... | | Authors: | Tholander, F, Haeggstrom, J, Thunnissen, M, Muroya, A, Roques, B.-P, Fournie-Zaluski, M.-C. | | Deposit date: | 2007-10-31 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.811 Å) | | Cite: | Structure-based dissection of the active site chemistry of leukotriene a4 hydrolase: implications for m1 aminopeptidases and inhibitor design.
Chem.Biol., 15, 2008
|
|
7W5Z
 
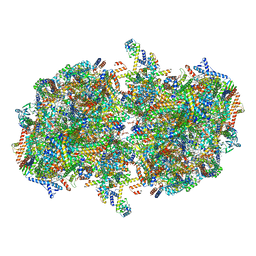 | | Cryo-EM structure of Tetrahymena thermophila mitochondrial complex IV, composite dimer model | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-oxoglutarate/malate carrier protein, CARDIOLIPIN, ... | | Authors: | Zhou, L, Maldonado, M, Padavannil, A, Letts, J. | | Deposit date: | 2021-11-30 | | Release date: | 2022-04-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structures of Tetrahymena 's respiratory chain reveal the diversity of eukaryotic core metabolism.
Science, 376, 2022
|
|
