3E0N
 
 | | The X-ray structure of Human Prostasin in complex with DFFR-chloromethyl ketone inhibitor | | Descriptor: | DPN-PHE-ARM, GLYCEROL, Prostasin heavy chain, ... | | Authors: | Spraggon, G, Hornsby, M, Shipway, A, Harris, J.L, Lesley, S.A. | | Deposit date: | 2008-07-31 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Active site conformational changes of prostasin provide a new mechanism of protease regulation by divalent cations.
Protein Sci., 18, 2009
|
|
3DOI
 
 | | Crystal Structure of a Thermostable Esterase complex with paraoxon | | Descriptor: | DIETHYL PHOSPHONATE, esterase | | Authors: | Levisson, M, Sun, L, Hendriks, S, Dijkstra, B.W, Van der Oost, J, Kengen, S.W.M. | | Deposit date: | 2008-07-04 | | Release date: | 2009-02-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure and biochemical properties of a novel thermostable esterase containing an immunoglobulin-like domain.
J.Mol.Biol., 385, 2009
|
|
3DPT
 
 | | COR domain of Rab family protein (Roco) | | Descriptor: | Rab family protein | | Authors: | Gotthardt, K, Weyand, M, Kortholt, A, Van Haastert, P.J.M, Wittinghofer, A. | | Deposit date: | 2008-07-09 | | Release date: | 2008-08-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the Roc-COR domain tandem of C. tepidum, a prokaryotic homologue of the human LRRK2 Parkinson kinase
Embo J., 27, 2008
|
|
3DKO
 
 | | Complex between the kinase domain of human ephrin type-a receptor 7 (epha7) and inhibitor alw-ii-49-7 | | Descriptor: | 5-[(2-methyl-5-{[3-(trifluoromethyl)phenyl]carbamoyl}phenyl)amino]pyridine-3-carboxamide, Ephrin type-A receptor 7 | | Authors: | Walker, J.R, Syeda, F, Gray, N, Butler-Cole, C, Bountra, C, Wolkstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-06-25 | | Release date: | 2008-08-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinase domain of human ephrin type-a receptor 7 (epha7) in complex with ALW-II-49-7
To be Published
|
|
7YFF
 
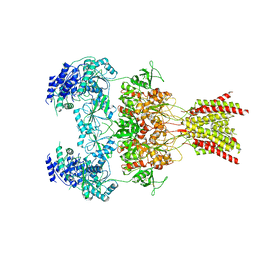 | | Structure of GluN1a-GluN2D NMDA receptor in complex with agonist glycine and competitive antagonist CPP. | | Descriptor: | (2R)-4-(3-phosphonopropyl)piperazine-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J.L, Zhu, S.J, Zhang, M. | | Deposit date: | 2022-07-08 | | Release date: | 2023-04-12 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7EEB
 
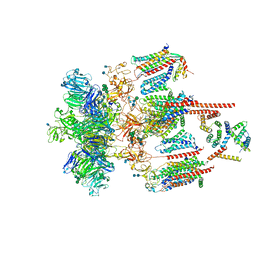 | | Structure of the CatSpermasome | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, J.P, Ke, M. | | Deposit date: | 2021-03-18 | | Release date: | 2021-07-28 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of a mammalian sperm cation channel complex.
Nature, 595, 2021
|
|
7YFL
 
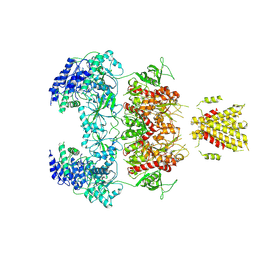 | | Structure of GluN1a-GluN2D NMDA receptor in complex with agonists glycine and glutamate. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, J.L, Zhu, S.J, Zhang, M. | | Deposit date: | 2022-07-08 | | Release date: | 2023-04-12 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7YFO
 
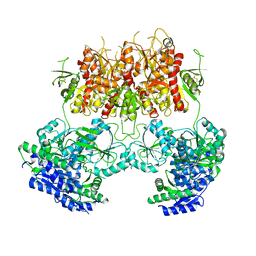 | |
7YFR
 
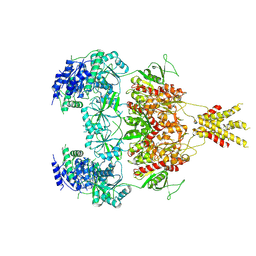 | | Structure of GluN1a E698C-GluN2D NMDA receptor in cystines non-crosslinked state. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, J.L, Zhu, S.J, Zhang, M. | | Deposit date: | 2022-07-09 | | Release date: | 2023-04-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
3DTE
 
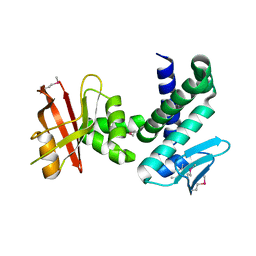 | | Crystal structure of the IRRE protein, a central regulator of DNA damage repair in deinococcaceae | | Descriptor: | IrrE protein | | Authors: | Vujicic-Zagar, A, Dulermo, R, Le Gorrec, M, Vannier, F, Servant, P, Sommer, S, De Groot, A, Serre, L. | | Deposit date: | 2008-07-15 | | Release date: | 2009-02-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the IrrE protein, a central regulator of DNA damage repair in deinococcaceae
J.Mol.Biol., 386, 2009
|
|
3DU2
 
 | |
7XNJ
 
 | |
3U52
 
 | |
7EIH
 
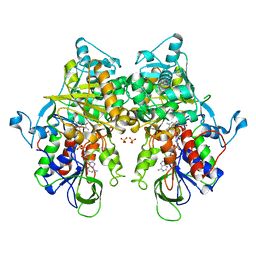 | | Ancestral L-Lys oxidase (ligand free form) | | Descriptor: | FAD dependent enzyme, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Sugiura, S, Nakano, S, Niwa, M, Hasebe, F, Ito, S. | | Deposit date: | 2021-03-31 | | Release date: | 2021-08-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Catalytic mechanism of ancestral L-lysine oxidase assigned by sequence data mining.
J.Biol.Chem., 297, 2021
|
|
7Y8N
 
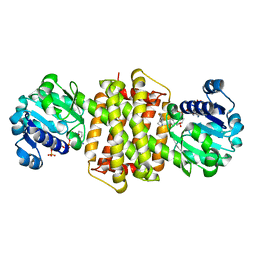 | | Structure of ScIRED-R3-V4 from Streptomyces clavuligerus in complex with 5-(2,5-difluorophenyl)-3,4-dihydro-2H-pyrrole | | Descriptor: | 5-[2,5-bis(fluoranyl)phenyl]-3,4-dihydro-2~{H}-pyrrole, Beta-hydroxyacid dehydrogenase, 3-hydroxyisobutyrate dehydrogenase, ... | | Authors: | Zhang, L.L, Liu, W.D, Shi, M, Huang, J.W, Yang, Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2022-06-24 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Engineered Imine Reductase for Larotrectinib Intermediate Manufacture
Acs Catalysis, 12, 2022
|
|
7EII
 
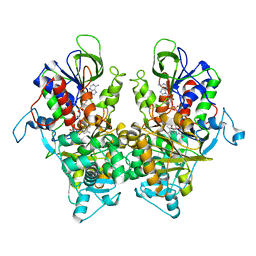 | | Ancestral L-Lys oxidase K387A variant (L-Lys binding form) | | Descriptor: | FAD dependent L-Lys oxidase, FLAVIN-ADENINE DINUCLEOTIDE, LYSINE | | Authors: | Sugiura, S, Nakano, S, Niwa, M, Hasebe, F, Ito, S. | | Deposit date: | 2021-03-31 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Catalytic mechanism of ancestral L-lysine oxidase assigned by sequence data mining.
J.Biol.Chem., 297, 2021
|
|
7FO8
 
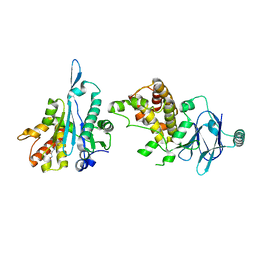 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P07H06 from the F2X-Universal Library | | Descriptor: | A1 cistron-splicing factor AAR2, N-[(1E)-2-(hydroxyamino)-2-oxoethylidene]benzamide, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7XS6
 
 | | structure of a membrane-integrated glycosyltransferase with inhibitor | | Descriptor: | (19R,22S)-25-amino-22-hydroxy-22-oxido-16-oxo-17,21,23-trioxa-22lambda~5~-phosphapentacosan-19-yl (9Z)-hexadec-9-enoate, (2S)-{[(2S,3S,4S)-2-amino-4-hydroxy-4-(5-hydroxypyridin-2-yl)-3-methylbutanoyl]amino}[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxyoxolan-2-yl]acetic acid (non-preferred name), Chitin synthase 1, ... | | Authors: | Wu, Y.N, Zhang, M, Yang, Y.Z, Ding, X.Y, Liu, X.T, Zhang, M.J, Yu, H.J. | | Deposit date: | 2022-05-12 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | structure of a membrane-integrated glycosyltransferase with inhibitor
To Be Published
|
|
7EIJ
 
 | | Ancestral L-Lys oxidase K387A variant (L-Arg binding form) | | Descriptor: | ARGININE, FAD dependent L-Lys oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Sugiura, S, Nakano, S, Niwa, M, Hasebe, F, Ito, S. | | Deposit date: | 2021-03-31 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Catalytic mechanism of ancestral L-lysine oxidase assigned by sequence data mining.
J.Biol.Chem., 297, 2021
|
|
3DKZ
 
 | | Crystal structure of the Q7W9W5_BORPA protein from Bordetella parapertussis. Northeast Structural Genomics Consortium target BpR208C. | | Descriptor: | Thioesterase superfamily protein | | Authors: | Vorobiev, S.M, Su, M, Seetharaman, J, Wang, D, Ciccosanti, C, Foote, E.L, Mao, L, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-06-26 | | Release date: | 2008-08-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the Q7W9W5_BORPA protein from Bordetella parapertussis.
To be Published
|
|
7XS7
 
 | | structure of a membrane-integrated glycosyltransferase | | Descriptor: | (19R,22S)-25-amino-22-hydroxy-22-oxido-16-oxo-17,21,23-trioxa-22lambda~5~-phosphapentacosan-19-yl (9Z)-hexadec-9-enoate, Chitin synthase 1, DODECANE, ... | | Authors: | Wu, Y.N, Zhang, M, Yang, Y.Z, Ding, X.Y, Liu, X.T, Zhang, M.J, Yu, H.J. | | Deposit date: | 2022-05-13 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | structure of a membrane-integrated glycosyltransferase with inhibitor
To Be Published
|
|
3DZD
 
 | | Crystal structure of sigma54 activator NTRC4 in the inactive state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, SODIUM ION, Transcriptional regulator (NtrC family) | | Authors: | Batchelor, J.D, Doucleff, M, Lee, C.-J, Matsubara, K, De Carlo, S, Heideker, J, Lamers, M.M, Pelton, J.G, Wemmer, D.E. | | Deposit date: | 2008-07-29 | | Release date: | 2008-11-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and regulatory mechanism of Aquifex aeolicus NtrC4: variability and evolution in bacterial transcriptional regulation.
J.Mol.Biol., 384, 2008
|
|
6MDD
 
 | |
7EIR
 
 | | Crystal structure of chondroitin ABC lyase I in complex with chondroitin disaccharide 6S | | Descriptor: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-6-O-sulfo-beta-D-galactopyranose, Chondroitin sulfate ABC endolyase, GLYCEROL, ... | | Authors: | Takashima, M, Miyanaga, A, Eguchi, T. | | Deposit date: | 2021-03-31 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Substrate specificity of Chondroitinase ABC I based on analyses of biochemical reactions and crystal structures in complex with disaccharides.
Glycobiology, 31, 2021
|
|
7EIP
 
 | | Crystal structure of ligand-free chondroitin ABC lyase I | | Descriptor: | ACETATE ION, Chondroitin sulfate ABC endolyase, MAGNESIUM ION | | Authors: | Takashima, M, Miyanaga, A, Eguchi, T. | | Deposit date: | 2021-03-31 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Substrate specificity of Chondroitinase ABC I based on analyses of biochemical reactions and crystal structures in complex with disaccharides.
Glycobiology, 31, 2021
|
|
